 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.SV2MR | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 366aa MW: 41654.9 Da PI: 8.595 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 59.7 | 6.3e-19 | 76 | 123 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd llv +++++G+g+Wk+I+++ g+ R+ k+c++rw +yl
Araip.SV2MR 76 KGPWTPEEDILLVSYIQEHGPGNWKAIPSKTGLLRCSKSCRLRWTNYL 123
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 48.5 | 2.1e-15 | 129 | 174 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T +E+ ++++ ++lG++ W++Ia++++ Rt++++k++w++yl
Araip.SV2MR 129 RGNFTNQEENMIIHLQALLGNR-WAAIASYLP-QRTDNDIKNYWNTYL 174
89********************.*********.*************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 8.7E-25 | 67 | 126 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 25.467 | 71 | 127 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.01E-30 | 73 | 170 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.6E-16 | 75 | 125 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.4E-17 | 76 | 123 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.49E-11 | 78 | 123 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-24 | 127 | 178 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.1E-15 | 128 | 176 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 19.956 | 128 | 178 | IPR017930 | Myb domain |
| Pfam | PF00249 | 1.1E-14 | 129 | 174 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.61E-10 | 131 | 174 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001666 | Biological Process | response to hypoxia | ||||
| GO:0009617 | Biological Process | response to bacterium | ||||
| GO:0009626 | Biological Process | plant-type hypersensitive response | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0042761 | Biological Process | very long-chain fatty acid biosynthetic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 366 aa Download sequence Send to blast |
VSVHFTHSIY NLIFNYLNSP IYTLTILSLC YAMLGLLHST TQIETNTIES STKKRNYYIF 60 VIMGRPPCCD KEGIKKGPWT PEEDILLVSY IQEHGPGNWK AIPSKTGLLR CSKSCRLRWT 120 NYLRPGIKRG NFTNQEENMI IHLQALLGNR WAAIASYLPQ RTDNDIKNYW NTYLKKRLSK 180 KLEEEKKDGS CLANNNNDFS SQKVVMPRGQ WERRLQTDIN MAKKALTHAL SPQNNNNNNI 240 NIESLSCSSS SNSNSNSCSS NNNNKATQSL SYASNADNIA RLLKGWMKKN NNNDTASSKE 300 GIFGTKVFGS EECLESSNNT NSSLEFSQSI YSQQEESKTE EIGEVLMPFH LLEKWLLDET 360 NIEKFI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 2e-25 | 76 | 178 | 27 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. {ECO:0000305}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00386 | DAP | Transfer from AT3G28910 | Download |
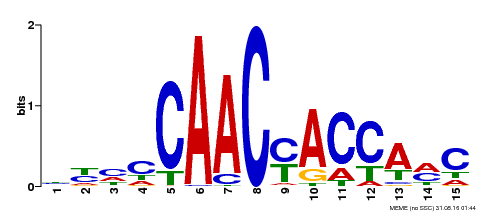
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.SV2MR |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016179105.1 | 0.0 | transcription factor MYB30-like | ||||
| Refseq | XP_025684174.1 | 0.0 | transcription factor MYB30 | ||||
| Swissprot | P81392 | 4e-99 | MYB06_ANTMA; Myb-related protein 306 | ||||
| TrEMBL | A0A444X8J3 | 0.0 | A0A444X8J3_ARAHY; Uncharacterized protein | ||||
| STRING | XP_007151227.1 | 1e-118 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1016 | 34 | 115 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G28910.1 | 6e-90 | myb domain protein 30 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



