 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.TUB1K | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 1058aa MW: 118872 Da PI: 5.4077 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43 | 1.1e-13 | 117 | 162 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
g W Ede+l+ av ++G+++W++I++ + ++++kqck rw+ +l
Araip.TUB1K 117 GVWKNTEDEILKAAVMKYGKNQWARISSLLV-RKSAKQCKARWYEWL 162
78*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 32.7 | 1.8e-10 | 169 | 212 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
WT eEde+l+++ k++++ W+tIa +g Rt+ qc +r+ k+l
Araip.TUB1K 169 TEWTREEDEKLLHLAKLMPTQ-WRTIAPIVG--RTPSQCLERYEKLL 212
68*****************99.********8..**********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 22.008 | 111 | 166 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 5.8E-18 | 114 | 165 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.0E-15 | 115 | 164 | IPR001005 | SANT/Myb domain |
| Pfam | PF13921 | 4.6E-13 | 119 | 179 | No hit | No description |
| CDD | cd00167 | 8.60E-12 | 119 | 162 | No hit | No description |
| SuperFamily | SSF46689 | 1.4E-20 | 142 | 215 | IPR009057 | Homeodomain-like |
| CDD | cd11659 | 1.65E-30 | 164 | 215 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-14 | 166 | 213 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.7E-12 | 167 | 214 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 16.443 | 167 | 216 | IPR017930 | Myb domain |
| Pfam | PF11831 | 3.8E-56 | 515 | 761 | IPR021786 | Pre-mRNA splicing factor component Cdc5p/Cef1 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009870 | Biological Process | defense response signaling pathway, resistance gene-dependent | ||||
| GO:0010204 | Biological Process | defense response signaling pathway, resistance gene-independent | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1058 aa Download sequence Send to blast |
MASPREPNDS HHRRYPSLVE SAVAADTLLL GWRCGTDRPV TIVPLLLFSL PSSSLFSSQS 60 PSPSSSLFSL PSPSISLFSS PSPSSSLSVA VELAVCFLSV SLSLESGGKM RIMIKGGVWK 120 NTEDEILKAA VMKYGKNQWA RISSLLVRKS AKQCKARWYE WLDPSIKKTE WTREEDEKLL 180 HLAKLMPTQW RTIAPIVGRT PSQCLERYEK LLDAACVKDE NYEPGDDPRK LRPGEIDPNP 240 ESKPARPDPV DMDEDEKEML SEARARLANT KGKKAKRKAR EKQLEEARRL ASLQKKRELK 300 AAGIDVRHRR RKRKGIDYNA EIPFEKRPPP GFFDIADEDR PVEQPQFPTT IEELEGKRRI 360 DVETQLRKQD IAKNKIAQRQ DAPSAILHAN KLNDPETVRK QTKLMLPPPQ ISDQELDEIA 420 RMGYANDLAG SDEFGEGSAA TRALLANYAQ TPGRGMTPLR IPQRSPASKG DAVMMEAENL 480 ARLRESQTPL LGGENPELHP SDFTGVTPKR KEVQTPNPML TPSATPGVAG LTPRIGMTPA 540 RDGYASMTPK GTPLRDELHI NEDVDMHERA KLELQRAEIR RSLRSGLGSL PQPKNEYQIV 600 MQPIPEDAEE PEEKIEEDMS DRIAREMAEE EARQQALLRK RSKVLQRELP RPPAASLELI 660 RNSLIRADGD KSSFVPPTSI EQADEMIRRE LLALLEHDNA KYPLDEKVNK EKKKGAKRVV 720 NEPVVPVIEE FHEEEMKDAD KLIKDEAHYL RVALGHESEP LEEFIEAHRT GLNDLMFFPT 780 RNAYGLSSVA GNMEKLAALQ NEFENVRKKF DDGKEKVVKL EKKAILLTQG YEAFRNAFCY 840 ARGYLHPIGV DKILEMRAKN SLWPQIEATS KQMDIAATEL ECFEALQKQE KLAASHRINN 900 LWEEVQKQKE LERTLQYKYG NLMEEVERIQ NVMEQYRVQA QKQEEIEANN RAHESTETVA 960 DKTGAQDSES FKAAPLTEEN ETAIATALSH DESEAGTVQD QTTASQKNDV DVDSDDIKQT 1020 EDPNAIPPDA ALAAKDDAGK VEGTITDGHI DNCKAALD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5mqf_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 5xjc_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 5yzg_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 5z56_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 5z57_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 5z58_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 6ff4_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 6ff7_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 6icz_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 6id0_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 6id1_L | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| 6qdv_O | 0.0 | 111 | 931 | 3 | 800 | Cell division cycle 5-like protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 294 | 313 | KKRELKAAGIDVRHRRRKRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Component of the MAC complex that probably regulates defense responses through transcriptional control and thereby is essential for plant innate immunity. Possesses a sequence specific DNA sequence 'CTCAGCG' binding activity. Involved in mRNA splicing and cell cycle control. May also play a role in the response to DNA damage. {ECO:0000250|UniProtKB:Q99459, ECO:0000269|PubMed:17298883, ECO:0000269|PubMed:17575050, ECO:0000269|PubMed:8917598}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00028 | SELEX | Transfer from AT1G09770 | Download |
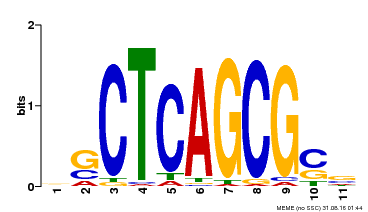
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.TUB1K |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016184908.1 | 0.0 | cell division cycle 5-like protein isoform X1 | ||||
| Swissprot | P92948 | 0.0 | CDC5L_ARATH; Cell division cycle 5-like protein | ||||
| TrEMBL | A0A445E775 | 0.0 | A0A445E775_ARAHY; Uncharacterized protein | ||||
| STRING | GLYMA10G30870.1 | 0.0 | (Glycine max) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4072 | 33 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09770.1 | 0.0 | cell division cycle 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



