 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Araip.ZP5MJ | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fabales; Fabaceae; Papilionoideae; Dalbergieae; Arachis
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 329aa MW: 37132.9 Da PI: 5.7458 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 46.8 | 7e-15 | 18 | 65 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEd++l ++ G g+W+ +ar g+ R++k+c++rw +yl
Araip.ZP5MJ 18 KGLWSPEEDDKLMNYMMNNGQGCWSDVARNAGLQRCGKSCRLRWINYL 65
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 47.4 | 4.4e-15 | 71 | 114 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg++++ E++l+++ + +lG++ W+ Ia++++ gRt++++k++w++
Araip.ZP5MJ 71 RGAFSPHEQQLIIHFHSLLGNR-WSQIAARLP-GRTDNEIKNFWNS 114
89********************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 6.8E-24 | 11 | 68 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.138 | 13 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.56E-27 | 16 | 112 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.5E-10 | 17 | 67 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 8.8E-14 | 18 | 65 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.46E-9 | 21 | 65 | No hit | No description |
| PROSITE profile | PS51294 | 23.823 | 66 | 120 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.6E-25 | 69 | 121 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.8E-14 | 70 | 118 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.5E-13 | 71 | 114 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.98E-10 | 73 | 113 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 329 aa Download sequence Send to blast |
MMITFEQSSS SKNNKLRKGL WSPEEDDKLM NYMMNNGQGC WSDVARNAGL QRCGKSCRLR 60 WINYLRPDLK RGAFSPHEQQ LIIHFHSLLG NRWSQIAARL PGRTDNEIKN FWNSTIKKRL 120 KQQSTTSPNA SDDHDYSSYD PNNNNNIVGG GEGFNSNSTT QQFHQHHLHL MPLVFNSSSS 180 SSSSSMQNTV LNTLIDRLPM LDHGLNIPAA NGFNNKGFYD LENGVIGSLD NNNNNNNINN 240 LVAEEVVVGD NTNTNTNTFS DHDNNLKVLE SANYFDDINS IVNSCNNNKI MKGGVVENLF 300 HEELTLGEWD LEEFMKDVTS FPFLADFSN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 1e-26 | 13 | 120 | 22 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00599 | PBM | Transfer from AT5G12870 | Download |
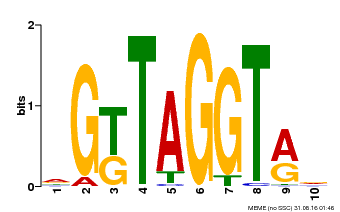
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Araip.ZP5MJ |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016193161.1 | 0.0 | myb-related protein 330 | ||||
| Refseq | XP_025642077.1 | 0.0 | transcription factor MYB83-like | ||||
| TrEMBL | A0A445A996 | 0.0 | A0A445A996_ARAHY; Uncharacterized protein | ||||
| STRING | XP_007132019.1 | 1e-122 | (Phaseolus vulgaris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF774 | 34 | 128 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G12870.1 | 2e-70 | myb domain protein 46 | ||||



