 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA001090-PA | ||||||||
| Common Name | OsI_03129 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 269aa MW: 28641.4 Da PI: 4.2221 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 63.9 | 3.5e-20 | 71 | 121 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.psengkr.krfslgkfgtaeeAakaaiaarkkleg 55
++++GVr+++ +g+++AeIrd + r r++lg+f+taeeAa+ +++a+ +l+g
BGIOSGA001090-PA 71 TKFRGVRRRP-WGKFAAEIRDpW-----RgVRVWLGTFDTAEEAARVYDNAAIQLRG 121
69********.**********88.....55*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.2E-12 | 71 | 121 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.17E-30 | 71 | 131 | No hit | No description |
| SMART | SM00380 | 7.8E-37 | 72 | 135 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 9.4E-30 | 72 | 130 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 6.08E-22 | 72 | 131 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 23.024 | 72 | 129 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.0E-10 | 73 | 84 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.0E-10 | 95 | 111 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042991 | Biological Process | transcription factor import into nucleus | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 269 aa Download sequence Send to blast |
MSRSRTVRIF WDDPDLTDSS GEDEGCGGRR VGSMCSGGDG DMGRRVVGGG CTVGVGRRRL 60 TKDGGPGAPS TKFRGVRRRP WGKFAAEIRD PWRGVRVWLG TFDTAEEAAR VYDNAAIQLR 120 GPSATTNFSA STNSAGAQDP VAVGYESGAE SSPAVSSPTS VLRKVPSLCS LAEDKDDYEA 180 GPCEPATAAG SNLTVLEEEE LGEFVPFEDA PVYGGSSFWD FEPESGFLYA EPSSPETPWD 240 AGATSSGEAQ DYFQDLRDLF PLNPLPAIF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 2e-22 | 65 | 133 | 7 | 76 | Ethylene-responsive transcription factor ERF096 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.38422 | 0.0 | callus| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 33440021 | 0.0 | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00456 | DAP | Transfer from AT4G27950 | Download |
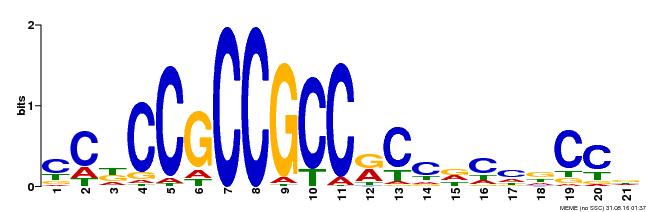
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP003294 | 0.0 | AP003294.2 Oryza sativa Japonica Group genomic DNA, chromosome 1, PAC clone:P0694A04. | |||
| GenBank | AP014957 | 0.0 | AP014957.1 Oryza sativa Japonica Group DNA, chromosome 1, cultivar: Nipponbare, complete sequence. | |||
| GenBank | AY339377 | 0.0 | AY339377.1 Oryza sativa (japonica cultivar-group) Ap24 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015635207.1 | 0.0 | pathogenesis-related genes transcriptional activator PTI6 | ||||
| TrEMBL | A2WTE5 | 0.0 | A2WTE5_ORYSI; Uncharacterized protein | ||||
| STRING | ORUFI01G28060.1 | 0.0 | (Oryza rufipogon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1690 | 36 | 97 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G11140.1 | 6e-25 | cytokinin response factor 1 | ||||



