 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA003304-PA | ||||||||
| Common Name | OsI_01468 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | M-type_MADS | ||||||||
| Protein Properties | Length: 277aa MW: 30116.6 Da PI: 8.3579 | ||||||||
| Description | M-type_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 56.9 | 2.7e-18 | 10 | 50 | 2 | 42 |
---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-T CS
SRF-TF 2 rienksnrqvtfskRrngilKKAeELSvLCdaevaviifss 42
ri n+ r tf kRr g+lKKA+EL +LCd++++++++++
BGIOSGA003304-PA 10 RIANDATRRATFKKRRRGLLKKASELATLCDVDACLVVYGE 50
899************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 19.105 | 1 | 50 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 1.5E-23 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00266 | 1.58E-34 | 2 | 85 | No hit | No description |
| SuperFamily | SSF55455 | 1.23E-24 | 3 | 92 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.0E-12 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 4.8E-18 | 10 | 50 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.0E-12 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.0E-12 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0043078 | Cellular Component | polar nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 277 aa Download sequence Send to blast |
MARKKIVLDR IANDATRRAT FKKRRRGLLK KASELATLCD VDACLVVYGE GDAEPEVWPS 60 TEVAMNVLRQ FRALPEMEQC KKMMNQEDFL RLRIGKLKEQ LRKMDRDNHE RETLILLHDA 120 LQGRLGTYES LSVEQLTSVD CLASARLKVI TDRLVEIRAP NEDGQVLVPP PPPPPPALPA 180 PPPPPAPMLP LAPPPTHVTP AMPLSSMPPP AFHGMNHHHH QNHFINHGGN DQNAWLMNVA 240 RNGGDLGALV YSAFASSSSS NTGGAGTSAA GAAAPGP |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 2 | 24 | RKKIVLDRIANDATRRATFKKRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00553 | DAP | Transfer from AT5G48670 | Download |
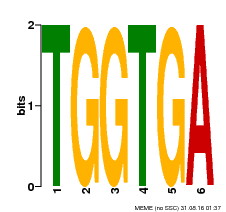
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012609 | 0.0 | CP012609.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 1 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025878343.1 | 0.0 | agamous-like MADS-box protein AGL80 | ||||
| TrEMBL | A2WNN8 | 0.0 | A2WNN8_ORYSI; Uncharacterized protein | ||||
| STRING | ORUFI01G13020.1 | 0.0 | (Oryza rufipogon) | ||||
| STRING | ONIVA01G14280.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP363 | 36 | 202 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G48670.1 | 3e-26 | AGAMOUS-like 80 | ||||



