 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA009504-PA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 794aa MW: 87091.8 Da PI: 5.6293 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 82.2 | 7e-26 | 1 | 53 | 26 | 78 |
HHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 26 vhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+h+ka++++v ++ qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
BGIOSGA009504-PA 1 MHAKATTAVVGNTVQRFCQQCSRFHPLQEFDEGKRSCRRRLAGHNRRRRKTRP 53
69************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03110 | 2.3E-18 | 1 | 50 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 21.786 | 1 | 51 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:4.10.1100.10 | 7.1E-16 | 1 | 38 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.19E-22 | 1 | 55 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 3.73E-8 | 574 | 681 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.0E-7 | 578 | 684 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 4.03E-8 | 581 | 681 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 794 aa Download sequence Send to blast |
MHAKATTAVV GNTVQRFCQQ CSRFHPLQEF DEGKRSCRRR LAGHNRRRRK TRPEVAVGGS 60 AFTEDKISSY LLLGLLGVCA NLNADNAEHL RGQELISGLL RNLGAVAKSL DPKELCKLLE 120 ACQSMQDGSN AGTSETANAL VNTAVAEAAG PSNSKMPFVN GDQCGLASSS VVPVQSKSPT 180 VATPDPPACK FKDFDLNDTY GGMEGFEDGY EGSPTPAFKT TDSPNCPSWM HQDSTQSPPQ 240 TSGNSDSTSA QSLSSSNGDA QCRTDKIVFK LFEKVPSDLP PVLRSQILGW LSSSPTDIES 300 YIRPGCIILT VYLRLVESAW KELSDNMSSY LDKLLNSSTG NFWASGLVFV MVRHQIAFMH 360 NGQLMLDRPL ANSAHHYCKI LCVRPIAAPF STKVNFRVEG LNLVSDSSRL ICSFEGSCIF 420 QEDTDNIVDD VEHDDIEYLN FCCPLPSSRG RGFVEVEDGG FSNGFFPFII AEQDICSEVC 480 ELESIFESSS HEQADDDNAR NQALEFLNEL GWLLHRANII SKQDKVPLAS FNIWRFRNLG 540 IFAMEREWCA VTKLLLDFLF TGLVDIGSQS PEEVVLSENL LHAAVRMKSA QMVRFLLGYK 600 PNESLKRTAE TFLFRPDAQG PSKFTPLHIA AATDDAEDVL DALTNDPGLV GINTWRNARD 660 GAGFTPEDYA RQRGNDAYLN MVEKKINKHL GKGHVVLGVP SSIHPVITDG VKPGEVSLEI 720 GMTVPPPAPS CNACSRQALM YPNSTARTFL YRPAMLTVMG IAVICVCVGL LLHTCPKVYA 780 APTFRWELLE RGPM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 1e-19 | 1 | 50 | 35 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 602 | 607 | ESLKRT |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.59197 | 0.0 | callus| flower| leaf| panicle| root| seed| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 32979871 | 0.0 | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Ubiquitous. {ECO:0000269|PubMed:16861571}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
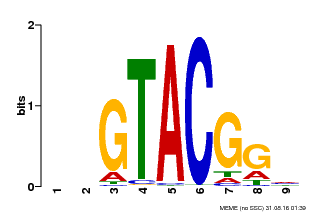
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK072164 | 0.0 | AK072164.1 Oryza sativa Japonica Group cDNA clone:J013130O11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631511.1 | 0.0 | squamosa promoter-binding-like protein 6 isoform X1 | ||||
| Swissprot | Q75LH6 | 0.0 | SPL6_ORYSJ; Squamosa promoter-binding-like protein 6 | ||||
| TrEMBL | A0A0P0W540 | 0.0 | A0A0P0W540_ORYSJ; Os03g0833300 protein (Fragment) | ||||
| STRING | ORUFI03G41710.1 | 0.0 | (Oryza rufipogon) | ||||
| STRING | OS03T0833300-02 | 0.0 | (Oryza sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1998 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



