 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA009880-PA | ||||||||
| Common Name | OsI_13373 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 357aa MW: 39338.2 Da PI: 6.6425 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 18.6 | 3.2e-06 | 289 | 318 | 26 | 55 |
-HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 26 saeereeLAkklgLterqVkvWFqNrRake 55
+++++ LA+++gL+ +q+ +WF N+R ++
BGIOSGA009880-PA 289 KESQKVALAESTGLDLKQINNWFINQRKRH 318
5788999********************885 PP
| |||||||
| 2 | ELK | 37.1 | 6.6e-13 | 218 | 239 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELKh+Ll+KYsgyL+sLkqE+s
BGIOSGA009880-PA 218 ELKHHLLKKYSGYLSSLKQELS 239
9*******************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01256 | 3.7E-28 | 129 | 180 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 1.0E-22 | 135 | 178 | IPR005541 | KNOX2 |
| SMART | SM01188 | 1800 | 138 | 158 | IPR005539 | ELK domain |
| SMART | SM01188 | 4.9E-7 | 218 | 239 | IPR005539 | ELK domain |
| Pfam | PF03789 | 8.1E-10 | 218 | 239 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 11.331 | 218 | 238 | IPR005539 | ELK domain |
| SMART | SM00389 | 6.7E-6 | 240 | 325 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 8.5E-26 | 243 | 322 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 2.13E-8 | 250 | 322 | No hit | No description |
| Pfam | PF05920 | 1.4E-9 | 258 | 317 | IPR008422 | Homeobox KN domain |
| PROSITE profile | PS50071 | 9.297 | 290 | 321 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.88E-9 | 290 | 334 | IPR009057 | Homeodomain-like |
| PROSITE pattern | PS00027 | 0 | 296 | 319 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 357 aa Download sequence Send to blast |
MEEISHHFGV VGASGVHGGH QHQHQHHHHP WGSSLSAIVA PPPPPQPQQQ QTQAGGMAHT 60 PLTLNTAAAA VGNPVLQLAN GSLLDACGKA KEASASASYA ADVGAPPEVA ARLTAVAQDL 120 ELRQRTALGV LGAATEPELD QFMEAYHEML VKYREELTRP LQEAMEFLRR VETQLNTLSI 180 SGRSLRNILS SGSSEEDQEG SGGETELPEI DAHGVDQELK HHLLKKYSGY LSSLKQELSK 240 KKKKGKLPKD ARQQLLNWWE LHYKWPYPSA YSTANTCDVC TVDSVISIKE SQKVALAEST 300 GLDLKQINNW FINQRKRHWK PSDEMQFVMM DGYHPTNAAA FYMDGHFIND GGLYRLG |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.151 | 0.0 | panicle| root| stem| vegetative meristem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 452235 | 0.0 | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the globular stage embryo 2 days after pollination (DAP) in a restricted small region just below the center of the ventral region of the embryo, where the shoot apex arises later. From 3 to 4 DAP expressed in an enlarged ventral region of embryo, corresponding to the expected epiblast and radicle, respectively. At the shoot apex differentiation stage (4-5 DAP), expressed in the shoot apex, epiblast, radicle primordia, and in their intervening tissues. Expression in the radicle is observed in the cells surrounding the root apical meristem in a donut shape but not in the meristem. During the first and second leaf primordium formation, expression pattern is maintained, but decreases. During inflorescence development, expressed only in the corpus of the rachis primordium but not in the tunica layer (L1). After floral induction, expressed in both tunica and corpus but not in floral organ primordia. Later in flower development, expressed in the corpus of the floral meristem. {ECO:0000269|PubMed:10488233, ECO:0000269|PubMed:8755613, ECO:0000269|PubMed:9869405}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed predominantly in shoot apices. Also found to a lesser extent in glumes. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that regulates genes involved in development. May be involved in shoot formation during embryogenesis. Overexpression in transgenic plants causes altered leaf morphology (PubMed:10488233, PubMed:8755613, PubMed:9869405). Regulates anther dehiscence via direct repression of the auxin biosynthetic gene YUCCA4 (PubMed:29915329). Binds to the DNA sequence 5'-TGAC-3' in the promoter of the YUCCA4 gene and represses its activity during anther development (PubMed:29915329). Reduction of auxin levels at late stage of anther development, after meiosis of microspore mother cells, is necessary for normal anther dehiscence and seed setting (PubMed:29915329). {ECO:0000269|PubMed:10488233, ECO:0000269|PubMed:29915329, ECO:0000269|PubMed:8755613, ECO:0000269|PubMed:9869405}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
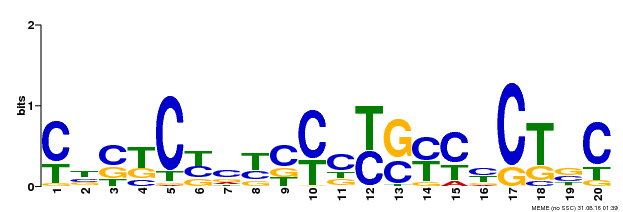
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FJ940206 | 0.0 | FJ940206.1 Oryza sativa Japonica Group clone KCB522D03 homeobox protein mRNA, complete cds. | |||
| GenBank | RICOSH1 | 0.0 | D16507.1 Oryza sativa Japonica Group OSH1 mRNA for homeobox protein, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015629392.1 | 0.0 | homeobox protein knotted-1-like 6 | ||||
| Swissprot | P46609 | 0.0 | KNOS6_ORYSJ; Homeobox protein knotted-1-like 6 | ||||
| TrEMBL | B8AR09 | 0.0 | B8AR09_ORYSI; Uncharacterized protein | ||||
| STRING | ONIVA03G34200.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9886 | 31 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 8e-74 | KNOTTED-like from Arabidopsis thaliana | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



