 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA016726-PA | ||||||||
| Common Name | OsI_16549 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 659aa MW: 67709.9 Da PI: 6.5076 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 44.7 | 3.3e-14 | 279 | 338 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pse.ng..kr.krfslgkfgtaeeAakaaiaarkkleg 55
s y+GV++++++gr++A+++d + g ++ ++++lg ++ +++Aa+a++ a++k++g
BGIOSGA016726-PA 279 SIYRGVTRHRWTGRYEAHLWDnSCRrEGqsRKgRQVYLGGYDKEDKAARAYDLAALKYWG 338
57*******************666664477446**********99*************98 PP
| |||||||
| 2 | AP2 | 51.2 | 3e-16 | 381 | 432 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+++++ grW A+I + +k ++lg+f t+eeAa+a++ a+ k++g
BGIOSGA016726-PA 381 SKYRGVTRHHQHGRWQARIGRVAG---NKDIYLGTFSTEEEAAEAYDIAAIKFRG 432
89****************988532...5************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 6.46E-19 | 279 | 348 | No hit | No description |
| Pfam | PF00847 | 2.8E-11 | 279 | 338 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.05E-16 | 279 | 348 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 1.7E-28 | 280 | 352 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 18.861 | 280 | 346 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 6.3E-15 | 280 | 348 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.3E-6 | 281 | 292 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 4.01E-23 | 381 | 442 | No hit | No description |
| Pfam | PF00847 | 1.7E-10 | 381 | 432 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.42E-18 | 381 | 442 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 4.3E-34 | 382 | 446 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.0E-18 | 382 | 441 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.401 | 382 | 440 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.3E-6 | 422 | 442 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000723 | Biological Process | telomere maintenance | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007389 | Biological Process | pattern specification process | ||||
| GO:0010449 | Biological Process | root meristem growth | ||||
| GO:0019827 | Biological Process | stem cell population maintenance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 659 aa Download sequence Send to blast |
MASADNWLGF SLSGQGNPQH HQNGSPSAAG DAAIDISGSG DFYGLPTPDA HHIGMAGEDA 60 PYGVMDAFNR GTHETQDWAM RGLDYGGGSS DLSMLVGSSG GGRRTVAGDG GGEAPKLENF 120 LDGNSFSDVH GQAAGGYLYS GSAVGGAGGY SNGGCGGGTI ELSMIKTWLR SNQSQQQPSP 180 PQHADQGMST DASASSYACS DVLVGSCGGG GGAGGTASSH GQGLALSMST GSVAAAAGGG 240 AVVAAESSSS ENKRVDSPGG AVDVAVPRKS IDTFGQRTSI YRGVTRHRWT GRYEAHLWDN 300 SCRREGQSRK GRQVYLGGYD KEDKAARAYD LAALKYWGTT TTTNFPMSNY EKELEEMKHM 360 TRQEYIAHLR RNSSGFSRGA SKYRGVTRHH QHGRWQARIG RVAGNKDIYL GTFSTEEEAA 420 EAYDIAAIKF RGLNAVTNFD MSRYDVKSIL DSSTLPVGGA ARRLKEAEAA AAAAGGGVIV 480 SHLADGGVGG YYYGCGPTIA FGGGGQQPAP LAVHYPSYGQ ASGWCKPEQD AVIAAGHCAT 540 DLQHLHLGSG GAAATHNFFQ QPASSSAVYG NGGGGGGNAF MMPMGAVVAA ADHGGQSSAY 600 GGGDESGRLV VGYDGVVDPY AAMRSAYELS QGSSSSSVSV AKAANGYPDN WSSPFNGMG |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.86735 | 0.0 | callus| leaf | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in embryo throughout embryogenesis. Also present in free nuclear endosperm, but disappears once endosperm cellularization begins. {ECO:0000269|PubMed:12172019}. | |||||
| Uniprot | TISSUE SPECIFICITY: Mostly expressed in developing seeds. {ECO:0000269|PubMed:12172019}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that promotes cell proliferation, differentiation and morphogenesis, especially during embryogenesis. {ECO:0000269|PubMed:12172019}. | |||||
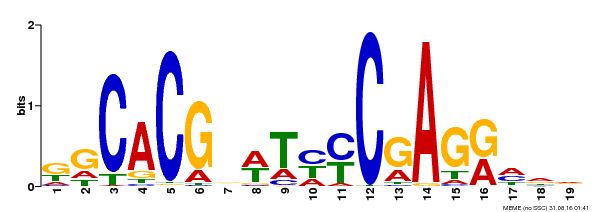
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00368 | DAP | Transfer from AT3G20840 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK287726 | 0.0 | AK287726.1 Oryza sativa Japonica Group cDNA, clone: J065138E17, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015635553.1 | 0.0 | AP2-like ethylene-responsive transcription factor BBM1 | ||||
| Swissprot | Q8LSN2 | 1e-135 | BBM2_BRANA; AP2-like ethylene-responsive transcription factor BBM2 | ||||
| TrEMBL | B8ARC2 | 0.0 | B8ARC2_ORYSI; Uncharacterized protein | ||||
| STRING | ORGLA04G0150400.1 | 0.0 | (Oryza glaberrima) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1569 | 37 | 110 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G20840.1 | 1e-120 | AP2 family protein | ||||



