 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA019856-PA | ||||||||
| Common Name | OsI_19936 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 934aa MW: 102801 Da PI: 6.5057 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.2 | 6.5e-35 | 168 | 242 | 2 | 76 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
Cqv+gCead+ e+k yhrrh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
BGIOSGA019856-PA 168 CQVPGCEADIRELKGYHRRHRVCLRCAHAAAVMLDGVQKRYCQQCGKFHILLDFDEDKRSCRRKLERHNKRRRRK 242
************************************************************************997 PP
| |||||||
| 2 | SBP | 40.8 | 6e-13 | 281 | 310 | 47 | 76 |
SSEEETTT--SS--S-STTTT-------S- CS
SBP 47 srfhelsefDeekrsCrrrLakhnerrrkk 76
srfh l +fDe+krsCrr+L++hn+rrr+k
BGIOSGA019856-PA 281 SRFHILLDFDEDKRSCRRKLERHNKRRRRK 310
9**************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.4E-27 | 162 | 229 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.487 | 165 | 242 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.75E-33 | 166 | 245 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 7.9E-27 | 168 | 242 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.09E-10 | 281 | 313 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.7E-6 | 281 | 310 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 10.802 | 281 | 310 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 934 aa Download sequence Send to blast |
MDAPGGGGGG GGGVDAGEPV WDWGNLLDFA VHDDDSLVLP WGDDSIGIEA DPAEAALLPP 60 APSPQPAEAE AEAAGPASLP SSMQAEGSKR RVRKRDPRLV CPNYLAGRVP CACPEIDEMA 120 AALEVEDVAT ELLAGARKKP KGAGRGSGAA VGGSGGGASR GTPAEMKCQV PGCEADIREL 180 KGYHRRHRVC LRCAHAAAVM LDGVQKRYCQ QCGKFHILLD FDEDKRSCRR KLERHNKRRR 240 RKPDSKGILE KDIDDQLDFS ADGSGDGELR EAIVQNSLPS SRFHILLDFD EDKRSCRRKL 300 ERHNKRRRRK PDSKGILEKD IDDQLDFSAD GSGDGELREE NIDVTTSETL ETVLSNKVLD 360 RETPVGSDDV LSSPTCAQPS LQIDQSKSLV TFAASVEACL GTKQENTKLT NSPVHDTKST 420 YSSSCPTGRV SFKLYDWNPA EFPRRLRHQI FEWLSSMPVE LEGYIRPGCT ILTVFVAMPQ 480 HMWDKLSEDT GNLVKSLVNA PNSLLLGKGA FFIHVNNMIF QVLKDGATLT STRLEVQSPR 540 IHYVHPSWFE AGKPIDLILC GSSLDQPKFR SLVSFDGLYL KHDCRRILSH ETFDCIGSGE 600 HILDSQHEIF RINITTSKLD THGPAFVEVE NMFGLSNFVP ILVGSKHLCS ELEQIHDALC 660 GSSDISSDPC ELRGLRQTAM LGFLIDIGWL IRKPSIDEFQ NLLSLANIQR WICMMKFLIQ 720 NDFINVLEII VNSLDNIIGS ELLSNLEKGR LENHVTEFLG YVSEARNIVD NRPKYDKQRQ 780 VDTRWAGDYA PNQPKLGISV PLAESTGTSG EHDLHTTNAA SGEEENMPLV TKALPHRQCC 840 HPETSARWLN AASIGAFPGG AMRMRLATTV VIGAVTTDSW TRLSSQIHCL KCLTNIESLK 900 RLSHSAAVKL IKAYRIVCCC AVLFPSKAVK QSSL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 4e-37 | 167 | 247 | 5 | 85 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 292 | 310 | KRSCRRKLERHNKRRRRKP |
| 2 | 296 | 308 | RRKLERHNKRRRR |
| 3 | 303 | 310 | NKRRRRKP |
| 4 | 304 | 309 | KRRRRK |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.19407 | 0.0 | callus| flower| leaf| panicle| root| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 32985266 | 0.0 | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Ubiquitous. {ECO:0000269|PubMed:16861571}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
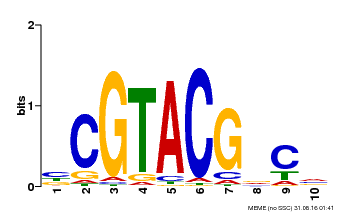
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK100057 | 0.0 | AK100057.1 Oryza sativa Japonica Group cDNA clone:J013158B16, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015640052.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | B8AY86 | 0.0 | B8AY86_ORYSI; Uncharacterized protein | ||||
| STRING | ONIVA05G16770.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-118 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



