 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA020778-PA | ||||||||
| Common Name | OsI_23911 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 344aa MW: 36899.5 Da PI: 9.3645 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 95.2 | 4.7e-30 | 191 | 250 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
++Dgy+WrKYGqK+ k++++pr+Y+rC++a Cpvkkkv+rsa+d++v++ tYegeHnh
BGIOSGA020778-PA 191 VKDGYQWRKYGQKVTKDNPCPRAYFRCSFApACPVKKKVQRSADDNTVLVATYEGEHNHA 250
58***************************99****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 26.199 | 186 | 252 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.3E-29 | 190 | 252 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.03E-25 | 190 | 252 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.2E-35 | 191 | 251 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 4.4E-25 | 192 | 249 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0002237 | Biological Process | response to molecule of bacterial origin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:1905034 | Biological Process | regulation of antifungal innate immune response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 344 aa Download sequence Send to blast |
MDSWIEQTSL SLDLNVGLPS TARRSSAPAA PIKVLVEENF LSFKKDHEVE ALEAELRRAS 60 EENKKLTEML RAVVAKYPEL QGQVNDMMSA AAAAAVNAGN HQSSTSEGGS VSPSRKRIRS 120 VDSLDDAAHH RKPSPPFVAA AAAAAYASPD QMECTSAAAA AAAKRVVRED CKPKVSKRFV 180 HADPSDLSLV VKDGYQWRKY GQKVTKDNPC PRAYFRCSFA PACPVKKKVQ RSADDNTVLV 240 ATYEGEHNHA QPPHHDAGSK TAAAAKHSQH QPPPSAAAAV VRQQQEQAAA AGPSTEVAAR 300 KNLAEQMAAT LTRDPGFKAA LVTALSGRIL ELSPTRTDPS LERR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 5e-22 | 190 | 253 | 13 | 75 | WRKY transcription factor 1 |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.50015 | 0.0 | callus| leaf| panicle| root| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 32991491 | 0.0 | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Regulates, probably indirectly, the activation of defense-related genes during defense response. Modulates plant innate immunity against X.oryzae pv. oryzae (Xoo). Regulates negatively the basal defense responses to the compatible fungus M.oryzae. {ECO:0000250|UniProtKB:Q6QHD1}. | |||||
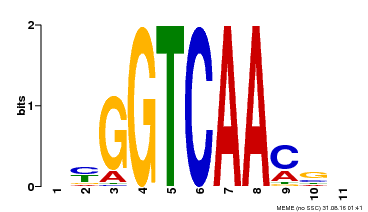
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00068 | PBM | Transfer from AT2G25000 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by biotic elicitors (e.g. fungal chitin oligosaccharide). Accumulates in response to M.oryzae. {ECO:0000250|UniProtKB:Q0DAJ3}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK106282 | 0.0 | AK106282.1 Oryza sativa Japonica Group cDNA clone:002-101-A04, full insert sequence. | |||
| GenBank | AK119644 | 0.0 | AK119644.1 Oryza sativa Japonica Group cDNA clone:002-130-G04, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015641578.1 | 0.0 | WRKY transcription factor WRKY28-like | ||||
| Swissprot | B8B0R8 | 0.0 | WRK28_ORYSI; WRKY transcription factor WRKY28 | ||||
| TrEMBL | A0A0E0HU98 | 0.0 | A0A0E0HU98_ORYNI; Uncharacterized protein | ||||
| STRING | OS06T0649000-01 | 0.0 | (Oryza sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1822 | 38 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G25000.1 | 7e-47 | WRKY DNA-binding protein 60 | ||||



