 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA021700-PA | ||||||||
| Common Name | OsI_22100 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 613aa MW: 67735 Da PI: 6.9184 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.9 | 5.3e-21 | 16 | 71 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
r++ +++t+ q+++Le++F+++++p++++r++L+++lgL+ rq+k+WFqNrR+++k
BGIOSGA021700-PA 16 RKRYHRHTPRQIQQLEAMFKECPHPDENQRAQLSRELGLEPRQIKFWFQNRRTQMK 71
688899***********************************************998 PP
| |||||||
| 2 | START | 157.4 | 1.2e-49 | 207 | 437 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE..........EXCCTTEEEEEEESSS.......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S. CS
START 2 laeeaaqelvkkalaeepgWvkss..........esengdevlqkfeeskv......dsgealrasgvvdmvlallveellddkeqWdetla. 77
+a +a++el+++a+a++ W+ks+ e +n d++ ++f++ ++ ++e +r+sg+v m ++ l ++d++ +W e ++
BGIOSGA021700-PA 207 MATRAMDELIRLAQAGDHIWSKSPgggvsggdarETLNVDTYDSIFSKPGGsyrapsINVEGSRESGLVLMSAVALADVFMDTN-KWMEFFPs 298
57899****************************999**********887779****99****************9999999999.******** PP
...EEEEEEEECTT.....EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCE CS
START 78 ...kaetlevissg.....galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksngh 161
ka t++v+ +g + l lm+ el ++p+vp R++ fvRy+rq ++g w+i+dvSvd +++ ++ R+++lpSg+li +++ng+
BGIOSGA021700-PA 299 ivsKAHTIDVLVNGmggrsESLILMYEELHIMTPAVPtREVNFVRYCRQIEQGLWAIADVSVDLQRDAHFGAPPPRSRRLPSGCLIADMANGY 391
*******************************************************************98899********************* PP
EEEEEEE-EE--SSXX.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 162 skvtwvehvdlkgrlp.hwllrslvksglaegaktwvatlqrqcek 206
skvtwveh+++++++p l+r lv sg+a+ga +w+a+lqr ce+
BGIOSGA021700-PA 392 SKVTWVEHMEVEEKSPiNVLYRDLVLSGAAFGAHRWLAALQRACER 437
****************999*************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.5E-20 | 9 | 76 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-22 | 10 | 78 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.204 | 13 | 73 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.8E-20 | 14 | 77 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 4.09E-20 | 15 | 74 | No hit | No description |
| Pfam | PF00046 | 1.5E-18 | 16 | 71 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 48 | 71 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 45.309 | 197 | 440 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 4.26E-31 | 198 | 439 | No hit | No description |
| CDD | cd08875 | 1.90E-112 | 201 | 436 | No hit | No description |
| SMART | SM00234 | 1.5E-35 | 206 | 437 | IPR002913 | START domain |
| Pfam | PF01852 | 6.4E-42 | 207 | 437 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.36E-12 | 454 | 568 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 613 aa Download sequence Send to blast |
MDFGDEPEGS DSQRRRKRYH RHTPRQIQQL EAMFKECPHP DENQRAQLSR ELGLEPRQIK 60 FWFQNRRTQM KAQHERADNC FLRAENDKIR CENIAIREAL KNVICPTCGG PPVGEDYFDE 120 QKLRMENARL KEELDRVSNL TSKYLGRPFT QLPPATPPMT VSSLDLSVGG MGGPSLDLDL 180 LSGGSSGIPF QLPAPVSDME RPMMAEMATR AMDELIRLAQ AGDHIWSKSP GGGVSGGDAR 240 ETLNVDTYDS IFSKPGGSYR APSINVEGSR ESGLVLMSAV ALADVFMDTN KWMEFFPSIV 300 SKAHTIDVLV NGMGGRSESL ILMYEELHIM TPAVPTREVN FVRYCRQIEQ GLWAIADVSV 360 DLQRDAHFGA PPPRSRRLPS GCLIADMANG YSKVTWVEHM EVEEKSPINV LYRDLVLSGA 420 AFGAHRWLAA LQRACERYAS LVALGVPHHI AGVTPEGKRS MMKLSQRMVN SFCSSLGASQ 480 MHQWTTLSGS NEVSVRVTMH RSTDPGQPNG VVLSAATSIW LPVPCDHVFA FVRDENTRSQ 540 WDVLSHGNQV QEVSRIPNGS NPGNCISLLR ILVSSLPSSK LNAESVATVN GLITTTVEQI 600 KAALNCSAHG HHP |
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 31339108 | 0.0 | ||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
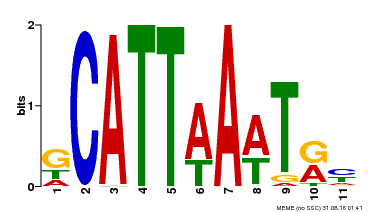
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP004758 | 0.0 | AP004758.3 Oryza sativa Japonica Group genomic DNA, chromosome 6, PAC clone:P0664C05. | |||
| GenBank | AP014962 | 0.0 | AP014962.1 Oryza sativa Japonica Group DNA, chromosome 6, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CP012614 | 0.0 | CP012614.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 6 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015693489.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein ROC8 | ||||
| Swissprot | Q69T58 | 0.0 | ROC8_ORYSJ; Homeobox-leucine zipper protein ROC8 | ||||
| TrEMBL | B8B3X5 | 0.0 | B8B3X5_ORYSI; Uncharacterized protein | ||||
| STRING | OBART06G06830.1 | 0.0 | (Oryza barthii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2912 | 37 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||



