 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA022077-PA | ||||||||
| Common Name | OsI_21349 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 345aa MW: 37825.2 Da PI: 10.2471 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 102.2 | 3.2e-32 | 178 | 231 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
BGIOSGA022077-PA 178 PRMRWTTTLHARFVHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLQMYRT 231
9****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.38E-15 | 175 | 232 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-28 | 176 | 231 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.5E-23 | 178 | 232 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.2E-7 | 179 | 230 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 345 aa Download sequence Send to blast |
MELFPRQLDL SLHISTPSCF SPSPASASTT ASSWPWTTKQ QLPEEEEAAA TTSTTTSSRS 60 APPLLLCNSD VFSTRGSSTT NTNHHADGGL ANSQQLKAAA RQPIHGIPVY HGHQQQQRRQ 120 LHHPYDVVGT RQSDGGRRLF SHHVGVGVTP SRTLSSSSSS PARLLPRLPP GRRSVRAPRM 180 RWTTTLHARF VHAVELLGGH ERATPKSVLE LMDVKDLTLA HVKSHLQMYR TIKNTDRPVS 240 NAGQNNDGFD NASAGDISDD SFTDGPLRQN KSMLASEQND TNIYSGLWSN NSSGKVGLPI 300 REPANEIYRR YLKNTKLSVD LETSMLRLPG RPNLEFTLGI GKASQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 3e-16 | 179 | 233 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 3e-16 | 179 | 233 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 3e-16 | 179 | 233 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 3e-16 | 179 | 233 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in developing phloem. {ECO:0000269|PubMed:14561401}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that regulates lateral organ polarity. Promotes abaxial cell fate during lateral organd formation. Functions with KAN1 in the specification of polarity of the ovule outer integument. {ECO:0000269|PubMed:11525739, ECO:0000269|PubMed:15286295, ECO:0000269|PubMed:16623911, ECO:0000269|PubMed:17307928}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
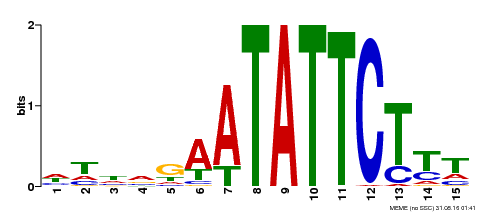
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012614 | 0.0 | CP012614.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 6 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006656560.1 | 1e-119 | PREDICTED: probable transcription factor KAN2 | ||||
| Swissprot | Q9C616 | 6e-48 | KAN2_ARATH; Probable transcription factor KAN2 | ||||
| TrEMBL | A2Y8H1 | 0.0 | A2Y8H1_ORYSI; Uncharacterized protein | ||||
| STRING | OMERI06G00740.1 | 0.0 | (Oryza meridionalis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP17435 | 11 | 11 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32240.1 | 2e-45 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



