 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA022875-PA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 448aa MW: 47849.8 Da PI: 4.7813 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 87.7 | 1.1e-27 | 173 | 227 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k++++WtpeLH+rFv+aveqL G++kA+P++ile+m++++Lt+++++SHLQkYR++
BGIOSGA022875-PA 173 KAKVDWTPELHRRFVQAVEQL-GIDKAVPSRILEIMGIDSLTRHNIASHLQKYRSH 227
6799*****************.********************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.924 | 170 | 229 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 6.45E-19 | 171 | 230 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-27 | 171 | 230 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.7E-26 | 173 | 227 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.1E-8 | 176 | 225 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0010638 | Biological Process | positive regulation of organelle organization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 448 aa Download sequence Send to blast |
MCPDIEDRAA VAGDAGMEVV GMSSDDMDQF DFSVDDIDFG DFFLRLEDGD VLPDLEVDPA 60 EIFTDFEAIA TSGGEGVQDQ EVPTVELLAP ADDVGVLDPC GDVVVGKENA AFAGAGEEKG 120 GCNQDDDAGE ANADDGAAAV EAKSSSPSSS TSSSQEAESR HKSSSKSSHG KKKAKVDWTP 180 ELHRRFVQAV EQLGIDKAVP SRILEIMGID SLTRHNIASH LQKYRSHRKH MIAREAEAAS 240 WTQRRQIYAA GGGAVAKRPE SNAWTVPTIG FPPPPPPPPS PAPMQHFARP LHVWGHPTMD 300 PSRVPVWPPR HLVPRGPAPP WVPPPPPSDP AFWHHPYMRG PAHVPTQGTP CMAMPMPAAR 360 FPAPPVPGVV PCPMYRPLTP PALTSKNQQD AQLQLQVQPS SESIDAAIGD VLSKPWLPLP 420 LGLKPPSVDS VMGELQRQGV ANVPPACG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 5e-17 | 170 | 225 | 2 | 57 | ARR10-B |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.3340 | 0.0 | callus| leaf| panicle| root| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 13940497 | 0.0 | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in leaves. {ECO:0000269|PubMed:11340194}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcriptional activator that promotes chloroplast development. Acts as an activator of nuclear photosynthetic genes involved in chlorophyll biosynthesis, light harvesting, and electron transport. {ECO:0000269|PubMed:19808806}. | |||||
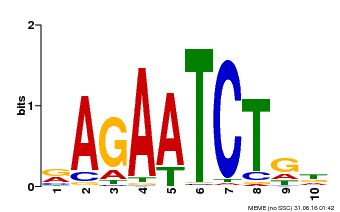
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By light. {ECO:0000269|PubMed:11340194}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF318581 | 0.0 | AF318581.1 Oryza sativa putative transcription factor OsGLK1 (Glk1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015644244.1 | 0.0 | probable transcription factor GLK1 | ||||
| Swissprot | Q5Z5I4 | 0.0 | GLK1_ORYSJ; Probable transcription factor GLK1 | ||||
| TrEMBL | Q93XP5 | 0.0 | Q93XP5_ORYSA; Putative transcription factor OsGLK1 | ||||
| STRING | ONIVA06G16920.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2415 | 38 | 86 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G20570.1 | 3e-48 | GBF's pro-rich region-interacting factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



