 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA023903-PA | ||||||||
| Common Name | OsI_26876 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 341aa MW: 37104.6 Da PI: 4.5305 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 61.2 | 2.3e-19 | 109 | 159 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
++y+G+r+++ +g+W+AeIrdp++ r++lg+fgtaeeAa a++ +++++g
BGIOSGA023903-PA 109 TRYRGIRQRP-WGKWAAEIRDPRK---GVRVWLGTFGTAEEAAMAYDVEARRIRG 159
68********.**********954...4************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 2.3E-12 | 109 | 159 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 2.16E-30 | 109 | 168 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 3.9E-31 | 110 | 168 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.18E-21 | 110 | 168 | IPR016177 | DNA-binding domain |
| PROSITE profile | PS51032 | 23.749 | 110 | 167 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.9E-35 | 110 | 173 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-11 | 111 | 122 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-11 | 133 | 149 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008219 | Biological Process | cell death | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051707 | Biological Process | response to other organism | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 341 aa Download sequence Send to blast |
MCGGSILSDL HLPVRRTVNA GDLWGDAGKG RDGGDGLKKR KGSSWDFDVD CDDDDDDFEA 60 DFEEFEDDYG DDDDVGFGHD DQESDMNGLK LAGFSTTKLG LGGSRKRKTR YRGIRQRPWG 120 KWAAEIRDPR KGVRVWLGTF GTAEEAAMAY DVEARRIRGK KAKVNFPDAA AAAPKRPRRS 180 SAKHSPQQQK ARSSSSSPAS LNASDAVSKS NNNRVSSAGS STDATAAAIA IDDGVKLELL 240 SETDPSPPMA AAAAAWLDAF ELNDLDGSRC KDNAFDHQIH KVEAAVADEF AFYDDPSYMQ 300 LGYQLDQGNS YENIDALFGG EAVNIGGLWS FDDMPMEFRA Y |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 2e-20 | 111 | 167 | 6 | 63 | ATERF1 |
| 3gcc_A | 2e-20 | 111 | 167 | 6 | 63 | ATERF1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 103 | 108 | SRKRKT |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.10293 | 0.0 | callus| flower| leaf| panicle| root| seed| stem | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 42821677 | 0.0 | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00109 | PBM | Transfer from AT3G16770 | Download |
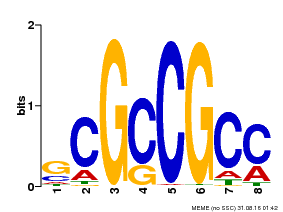
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK061047 | 0.0 | AK061047.1 Oryza sativa Japonica Group cDNA clone:006-205-F01, full insert sequence. | |||
| GenBank | AK067060 | 0.0 | AK067060.1 Oryza sativa Japonica Group cDNA clone:J013090P08, full insert sequence. | |||
| GenBank | AK109360 | 0.0 | AK109360.2 Oryza sativa Japonica Group cDNA clone:006-206-B03, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015647923.1 | 0.0 | ethylene-responsive transcription factor 1-like | ||||
| TrEMBL | A0A0E0I3P3 | 0.0 | A0A0E0I3P3_ORYNI; Uncharacterized protein | ||||
| TrEMBL | A2YNQ4 | 0.0 | A2YNQ4_ORYSI; Uncharacterized protein | ||||
| STRING | ONIVA07G20780.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP14560 | 21 | 23 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G16770.1 | 1e-33 | ethylene-responsive element binding protein | ||||



