 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | BGIOSGA029388-PA | ||||||||
| Common Name | OsI_32143 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza; Oryza sativa
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 315aa MW: 32021.5 Da PI: 8.754 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 228.2 | 1.5e-70 | 92 | 242 | 3 | 153 |
DUF702 3 sgtasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasaaeaaskrkrelkskkqsalsstklssae 95
g++sCqdCGnqakkdC+h+RCRtCCksrgf CathvkstWvpaakrrerqqqlaa++++aaa+a a + r+ +++ +++ ++++ ++ss++
BGIOSGA029388-PA 92 GGGISCQDCGNQAKKDCTHMRCRTCCKSRGFACATHVKSTWVPAAKRRERQQQLAALAASAAATAGGAGPSRDPTKRPRARPSATTPTTSSGD 184
5789********************************************************99999877777777778889999999999**** PP
DUF702 96 skkeletsslPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGle 153
+++ + ++++P+evsseavfrcvr++ vd++e+e+aYqtavsigGhvfkGiL+d G+e
BGIOSGA029388-PA 185 QQMVTVAERFPREVSSEAVFRCVRLGPVDQAEAEVAYQTAVSIGGHVFKGILHDVGPE 242
********************************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 5.8E-66 | 94 | 242 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01623 | 5.4E-27 | 96 | 138 | IPR006510 | Zinc finger, lateral root primordium type 1 |
| TIGRFAMs | TIGR01624 | 2.1E-22 | 193 | 241 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 315 aa Download sequence Send to blast |
MAGFPLGGGS HSRDNPAPPV PPVHPADAAS FLYATRGGSF QLWQQQEQQP FYASNIIRFA 60 DDAPPAPSLA GASSSSSSRG MRSSGGGGGG GGGGISCQDC GNQAKKDCTH MRCRTCCKSR 120 GFACATHVKS TWVPAAKRRE RQQQLAALAA SAAATAGGAG PSRDPTKRPR ARPSATTPTT 180 SSGDQQMVTV AERFPREVSS EAVFRCVRLG PVDQAEAEVA YQTAVSIGGH VFKGILHDVG 240 PEALAVAGGG GASEYHFRLT GDGSSPSTAA AGEAGSGGGG NIIVSSAVVM DPYPTPGPYG 300 AFPAGTPFFH GHPRP |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Os.8964 | 0.0 | callus| panicle| root | ||||
| Expression -- Microarray ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | ID | E-value | ||||
| GEO | 297609843 | 0.0 | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
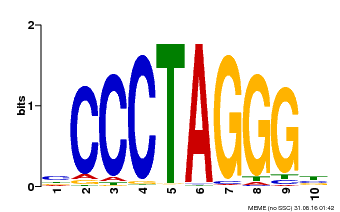
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK241285 | 0.0 | AK241285.1 Oryza sativa Japonica Group cDNA, clone: J065137A19, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015611315.1 | 0.0 | protein SHI RELATED SEQUENCE 1 | ||||
| TrEMBL | A0A0E0IMR7 | 0.0 | A0A0E0IMR7_ORYNI; Uncharacterized protein | ||||
| TrEMBL | A0A0E0QU83 | 0.0 | A0A0E0QU83_ORYRU; Uncharacterized protein | ||||
| TrEMBL | A2Z3E0 | 0.0 | A2Z3E0_ORYSI; Uncharacterized protein | ||||
| TrEMBL | Q652K4 | 0.0 | Q652K4_ORYSJ; Os09g0531600 protein | ||||
| STRING | ORUFI09G18770.1 | 0.0 | (Oryza rufipogon) | ||||
| STRING | OS09T0531600-01 | 0.0 | (Oryza sativa) | ||||
| STRING | ONIVA09G18410.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1354 | 35 | 124 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G66350.1 | 9e-41 | Lateral root primordium (LRP) protein-related | ||||



