 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bathy01g07090 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; prasinophytes; Mamiellophyceae; Mamiellales; Bathycoccaceae; Bathycoccus
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 757aa MW: 83891.4 Da PI: 6.1119 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 36.7 | 1e-11 | 198 | 242 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rWT E+ l+ d+ k++G Wk+ +++ ++t q++s+ qk+
Bathy01g07090 198 RERWTDAEHALFTDGLKMYGRA-WKKLEERVR-TKTVVQIRSHAQKF 242
78******************88.*********.************98 PP
| |||||||
| 2 | E2F_TDP | 69.5 | 4.1e-22 | 376 | 441 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
r++ksL ll+++fl+l+++ e+ ++l+ev+++L v +rRRiYDi+NVLea++++ kk kn++ w g
Bathy01g07090 376 RQSKSLSLLCERFLSLYSSGYENLISLDEVCSTL---GV--ERRRIYDIVNVLEAVEVVVKKGKNQYAWFG 441
679*******************************...**..****************************76 PP
| |||||||
| 3 | E2F_TDP | 59.3 | 5.9e-19 | 520 | 609 | 1 | 70 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvse...............dvknkrRRiYDilNVLealnliek.....kekneirwk 70
r+eksL l+tqkf++l+ ++e+g++ l+++a +++ + k+k RR+YDi+N+L++l+l++k ++k+++rw+
Bathy01g07090 520 RREKSLSLMTQKFITLFMEAEDGVLGLEDAAAAMLMSegstgpkatkdfndnELKKKIRRLYDIANILSSLRLLSKihlmdSRKPAFRWM 609
689*************************9999887666*************99999************************99*******6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.326 | 193 | 247 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.94E-13 | 193 | 248 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.2E-11 | 197 | 245 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.4E-8 | 197 | 245 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.9E-8 | 198 | 241 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.4E-5 | 198 | 239 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.87E-5 | 200 | 243 | No hit | No description |
| Gene3D | G3DSA:1.10.10.10 | 3.1E-24 | 372 | 442 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.09E-14 | 376 | 439 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 7.5E-26 | 376 | 441 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 9.8E-20 | 378 | 441 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 1.2E-18 | 516 | 609 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 4.9E-8 | 520 | 597 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 7.5E-23 | 520 | 610 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 5.7E-16 | 522 | 609 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032876 | Biological Process | negative regulation of DNA endoreduplication | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 757 aa Download sequence Send to blast |
MMMLLERKAT TTTRDDDDDI KNAAAAEEVS RENFQNHEGE DEKNTTTQEK KLKKKKKKKR 60 DVDGTTTMRD AEQGDDDQFV RGAEEERVAA KRKRKEEGEV VMKGLREGFG VNTTVATAET 120 TTTQTTTTTR EQRVEEEKEE EEEEEDERQP RDPMTIDCVD IEHTRAEDAP LSSSSSPPPL 180 LLLASSKKTT SATVKQPRER WTDAEHALFT DGLKMYGRAW KKLEERVRTK TVVQIRSHAQ 240 KFFDKLQRGE GMEEESKEEK KALLKAWARK VVGGGAGETS DTNTAGDVPA STPTTKATKK 300 KSGALTAEEA PPLSQVETAA TRMEAPTKRD ELNESIQKDV KDPMTTPTPT TEKFAAEVVD 360 YNIDGNTTSN SNKKTRQSKS LSLLCERFLS LYSSGYENLI SLDEVCSTLG VERRRIYDIV 420 NVLEAVEVVV KKGKNQYAWF GVSRLPSAIE KIEKFGAESF DIKLPEKLSL QVFENSLPFQ 480 SGGKDAINGV KSVANGEEDP TLPANKAKAT AAGKKETSER REKSLSLMTQ KFITLFMEAE 540 DGVLGLEDAA AAMLMSEGST GPKATKDFND NELKKKIRRL YDIANILSSL RLLSKIHLMD 600 SRKPAFRWMR AEDTIRKLIA TGKEHEWFGT TPKSPDARGQ LVVSPVKEGV SDDAAIGALT 660 TDTVPRLLPS AITALGRKDR HVFAALQTME KLLQAQAEDL SAKSAAQSTV PDGQKKIPEM 720 FLTTANLFGA GESFQSKTGD GIFNQYINNF TQNMRK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4yo2_A | 3e-32 | 373 | 602 | 1 | 228 | Transcription factor E2F8 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 49 | 57 | KKLKKKKKK |
| 2 | 49 | 58 | KKLKKKKKKK |
| 3 | 50 | 57 | KLKKKKKK |
| 4 | 53 | 59 | KKKKKKR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00397 | DAP | Transfer from AT3G48160 | Download |
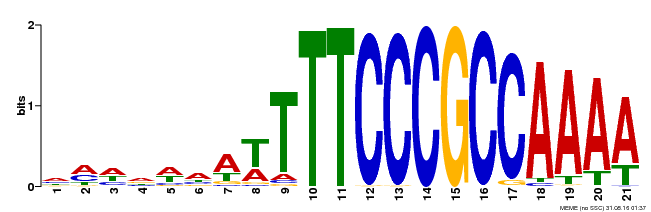
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FO082278 | 0.0 | FO082278.1 Bathycoccus prasinos genomic : Chromosome_1. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_007515521.1 | 0.0 | predicted protein | ||||
| TrEMBL | K8EPS5 | 0.0 | K8EPS5_9CHLO; Uncharacterized protein | ||||
| STRING | XP_007515521.1 | 0.0 | (Bathycoccus prasinos) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP4186 | 10 | 12 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48160.1 | 3e-35 | DP-E2F-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 19018443 |



