 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.0556s0021.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | BBR-BPC | ||||||||
| Protein Properties | Length: 286aa MW: 32007.5 Da PI: 10.3387 | ||||||||
| Description | BBR-BPC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GAGA_bind | 360.3 | 4e-110 | 1 | 285 | 1 | 301 |
GAGA_bind 1 mdddgsrernkgyyepa.aslkenlglqlmssiaerdaki....rernlalsekkaavaerd......maflqrdkalaernkalverdn 79
mdddg rn+gyy++a a++k nlglqlms+i +r++k+ re+nl ++++++++++r+ m++ +++++ +dn
Bostr.0556s0021.1.p 1 MDDDGL--RNWGYYDQAaATFKGNLGLQLMSTI-DRNTKPflpgRESNL-MIGSNGSYHPREheasmpMNY----SWINQ------SKDN 76
999999..9******9999**************.9**************.************998888888....*****......9*** PP
GAGA_bind 80 kllalllvenslasalpvgvqvlsgtksidslqqlsepqledsavelreeeklealpi.eeaaeeakekkkkkkrqrakkpkekkakkkk 168
k++++l++++ p++++vls+t+ +s+q++++p ++ s +e e+ + +p e ++ +k++k++ ++ ++++pk+kk +k+k
Bostr.0556s0021.1.p 77 KFFNMLPIST------PNYSNVLSETSGSNSMQMMHHPVVNSSRFE---EKPIPPPPReDEIVQPNKKRKMRGSTTTSTTPKAKKLRKPK 157
*******766......99*******************988888887...33333333303445556666777777788888888888888 PP
GAGA_bind 169 kksekskkkvkkesaderskaekksidlvlngvslDestlPvPvCsCtGalrqCYkWGnGGWqSaCCtttiSvyPLPvstkrrgaRiagr 258
+ +++v+++ +++r k kks+d+v+ngvs+D+s+lPvPvC+CtG+++qCY+WG+GGWqSaCCtt+iSvyPLP+stkrrgaRi+gr
Bostr.0556s0021.1.p 158 E-----ERDVTNNVQQQRLKPVKKSVDVVINGVSMDISGLPVPVCTCTGTPQQCYRWGCGGWQSACCTTNISVYPLPMSTKRRGARISGR 242
8.....4566666667889*********************************************************************** PP
GAGA_bind 259 KmSqgafkklLekLaaeGydlsnpvDLkdhWAkHGtnkfvtir 301
KmSqgafkk+LekLa+eGy+++n++DLk+hWA+HGtnkfvtir
Bostr.0556s0021.1.p 243 KMSQGAFKKVLEKLATEGYSFGNAIDLKSHWARHGTNKFVTIR 285
******************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01226 | 2.0E-167 | 1 | 285 | IPR010409 | GAGA-binding transcriptional activator |
| Pfam | PF06217 | 5.4E-101 | 1 | 285 | IPR010409 | GAGA-binding transcriptional activator |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0050793 | Biological Process | regulation of developmental process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 286 aa Download sequence Send to blast |
MDDDGLRNWG YYDQAAATFK GNLGLQLMST IDRNTKPFLP GRESNLMIGS NGSYHPREHE 60 ASMPMNYSWI NQSKDNKFFN MLPISTPNYS NVLSETSGSN SMQMMHHPVV NSSRFEEKPI 120 PPPPREDEIV QPNKKRKMRG STTTSTTPKA KKLRKPKEER DVTNNVQQQR LKPVKKSVDV 180 VINGVSMDIS GLPVPVCTCT GTPQQCYRWG CGGWQSACCT TNISVYPLPM STKRRGARIS 240 GRKMSQGAFK KVLEKLATEG YSFGNAIDLK SHWARHGTNK FVTIR* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional regulator that specifically binds to GA-rich elements (GAGA-repeats) present in regulatory sequences of genes involved in developmental processes. Negatively regulates the homeotic gene AGL11/STK, which controls ovule primordium identity, by a cooperative binding to purine-rich elements present in the regulatory sequence leading to DNA conformational changes. {ECO:0000269|PubMed:14731261, ECO:0000269|PubMed:15722463}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00253 | DAP | Transfer from AT2G01930 | Download |
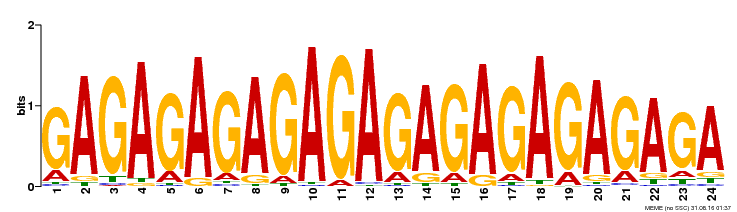
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.0556s0021.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC254589 | 0.0 | AC254589.1 Capsella rubella clone CAP16-D05, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020880017.1 | 0.0 | protein BASIC PENTACYSTEINE1 | ||||
| Refseq | XP_020880018.1 | 0.0 | protein BASIC PENTACYSTEINE1 | ||||
| Swissprot | Q9SKD0 | 1e-180 | BPC1_ARATH; Protein BASIC PENTACYSTEINE1 | ||||
| TrEMBL | D7LPX4 | 0.0 | D7LPX4_ARALL; Basic pentacysteine 1 | ||||
| STRING | Bostr.0556s0021.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2893 | 27 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01930.2 | 1e-180 | basic pentacysteine1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.0556s0021.1.p |



