 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.10064s0036.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 796aa MW: 86834 Da PI: 6.3187 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.6 | 6.8e-21 | 132 | 187 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++eLe++F+++++p++++r eL+k+l L++rqVk+WFqNrR+++k
Bostr.10064s0036.1.p 132 KKRYHRHTPQQIQELESMFKECPHPDEKQRLELSKRLCLETRQVKFWFQNRRTQMK 187
688999***********************************************999 PP
| |||||||
| 2 | START | 210.9 | 4.8e-66 | 318 | 537 | 1 | 206 |
HHHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS...SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EE CS
START 1 elaeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv..dsgealrasgvvdmvlallveellddkeqWdetla....ka 79
ela++a++elvk+a +eep+Wvks +++n+de++++f+++k +ea+r+sg+v+ ++ lve+l+d++ +W+e+++ +a
Bostr.10064s0036.1.p 318 ELALTAMDELVKLAHSEEPLWVKSLdgerDELNQDEYMRTFSSTKPtgLATEASRTSGMVIINSLALVETLMDSN-RWTEMFPcnvaRA 405
5899*************************************9999999***************************.************* PP
EEEEEECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCE CS
START 80 etlevissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksngh 161
+t++ is g galqlm+aelq+lsplvp R++ f+R+++q+ +g+w++vdvS+++ +++ s+v+R +lpSg++++++sng+
Bostr.10064s0036.1.p 406 TTTDIISGGmagtinGALQLMNAELQVLSPLVPvRNVNFLRFCKQHAEGVWAVVDVSIEPVRENAGGSPVIR--RLPSGCVVQDMSNGY 492
****************************************************************99******..*************** PP
EEEEEEE-EE--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 162 skvtwvehvdlkgrlphwllrslvksglaegaktwvatlqrqcek 206
skvtwveh++++++++h+l+r+l++sgl +g ++w+atlqrqce+
Bostr.10064s0036.1.p 493 SKVTWVEHAEYDENQIHQLYRPLLRSGLGFGSQRWLATLQRQCEC 537
*******************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.67E-20 | 119 | 189 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 6.8E-22 | 120 | 189 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.297 | 129 | 189 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 9.5E-18 | 130 | 193 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.49E-18 | 131 | 189 | No hit | No description |
| Pfam | PF00046 | 2.0E-18 | 132 | 187 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 164 | 187 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 42.074 | 309 | 540 | IPR002913 | START domain |
| CDD | cd08875 | 7.65E-112 | 313 | 536 | No hit | No description |
| SuperFamily | SSF55961 | 3.3E-29 | 313 | 537 | No hit | No description |
| SMART | SM00234 | 9.7E-39 | 318 | 537 | IPR002913 | START domain |
| Pfam | PF01852 | 2.1E-59 | 318 | 537 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 7.38E-21 | 556 | 780 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009827 | Biological Process | plant-type cell wall modification | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0043481 | Biological Process | anthocyanin accumulation in tissues in response to UV light | ||||
| GO:0048765 | Biological Process | root hair cell differentiation | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 796 aa Download sequence Send to blast |
MNFGSLFDNT PGGGSTGARL LSGLSYGNHT AATNNVLPGG AMAQAAGLFS PPLTKSVYAS 60 SGLSLALEQP ERRTNRGEAS MRKNNNNVGG DNFDGSVNRR SREDEHESRS GSDNVEGISG 120 EDQDAADKPP RKKRYHRHTP QQIQELESMF KECPHPDEKQ RLELSKRLCL ETRQVKFWFQ 180 NRRTQMKTQL ERHENALLRQ ENDKLRAENM SIREAMRNPI CTNCGGPAML GDVSLEEHHL 240 RIENARLKDE LDRVCNLTGK FLGHHHHHYN SSLELAVGTN NGGNFVFPPD FGGGACLPPP 300 QQQQTGINGI DQKSVLLELA LTAMDELVKL AHSEEPLWVK SLDGERDELN QDEYMRTFSS 360 TKPTGLATEA SRTSGMVIIN SLALVETLMD SNRWTEMFPC NVARATTTDI ISGGMAGTIN 420 GALQLMNAEL QVLSPLVPVR NVNFLRFCKQ HAEGVWAVVD VSIEPVRENA GGSPVIRRLP 480 SGCVVQDMSN GYSKVTWVEH AEYDENQIHQ LYRPLLRSGL GFGSQRWLAT LQRQCECLAV 540 LMSSSVTSHD NTSITPGGRK SMLKLAQRMT FNFCSGISAP SVHSWSKLTV GNVDPDVRVM 600 TRKSVDDPGE PPGIVLSAAT SVWLPASPQR LFDFLRNERI RCEWDILSNG GPMQEMAHIA 660 KGQDQGVSLL RSNAMNANQS SMLILQETCI DASGALVVYA PVDIPAMHVV MNGGDSSYVA 720 LLPSGFAVLP DGGLDGGSGD GEQRPVGGGS LLTVAFQILV NNLPTAKLTV ESVETVNNLI 780 SCTVQKIRAA LQCES* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the regulation of the tissue-specific accumulation of anthocyanins and in cellular organization of the primary root. {ECO:0000269|PubMed:10402424}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00421 | DAP | Transfer from AT4G00730 | Download |
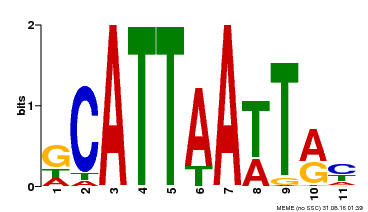
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.10064s0036.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK226968 | 0.0 | AK226968.1 Arabidopsis thaliana mRNA for homeodomain protein AHDP, complete cds, clone: RAFL09-36-B10. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006287095.1 | 0.0 | homeobox-leucine zipper protein ANTHOCYANINLESS 2 isoform X1 | ||||
| Swissprot | Q0WV12 | 0.0 | ANL2_ARATH; Homeobox-leucine zipper protein ANTHOCYANINLESS 2 | ||||
| TrEMBL | R0H8W0 | 0.0 | R0H8W0_9BRAS; Uncharacterized protein | ||||
| STRING | Bostr.10064s0036.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1128 | 27 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G00730.1 | 0.0 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.10064s0036.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



