 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.12659s0060.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 385aa MW: 43729 Da PI: 5.7492 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 30.1 | 1.1e-09 | 97 | 139 | 4 | 46 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLk 46
+k rr+++NReAAr+sR+RKka +++Lee +L++ ++L
Bostr.12659s0060.1.p 97 DKMKRRLAQNREAARKSRLRKKAHVQQLEESRLKLSQLEQELV 139
7899**************************8888887776665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 2.2E-7 | 88 | 140 | No hit | No description |
| SMART | SM00338 | 3.2E-6 | 90 | 176 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.347 | 96 | 138 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.9E-6 | 97 | 138 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 2.01E-6 | 98 | 140 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 101 | 116 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 1.6E-28 | 185 | 259 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009862 | Biological Process | systemic acquired resistance, salicylic acid mediated signaling pathway | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 385 aa Download sequence Send to blast |
MEMMSSSSSS TQVVSFRDMG MYEPFQQLSG WENPFKSDVN NISNNQNNNQ SSSTTLEVDA 60 RPEADDNNRA NYTSLYNNSV EAEPSSNNDQ DEDRINDKMK RRLAQNREAA RKSRLRKKAH 120 VQQLEESRLK LSQLEQELVK ARQQGLCVRS SSDTSYLGPA GNMNSGIAAF EMEYTHWLEE 180 QNRRVSEIRT ALQAHISDIE LKMLVDICLN HYANLFRMKA DAAKADVFFL MSGMWRTSTE 240 RFFQWIGGFR PSELLNVVMP YVEPLTNQQL LEVRNLQQSS QQAEEALSQG LDKLQQGLVE 300 SIAIQITVVE SVNHGAPMAS AMENLQALES FVNQADHLRQ QTLQQMSKIL TTRQAARGLL 360 ALGEYFYRLR ALSSLWAARP REQT* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
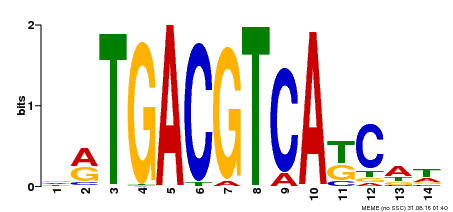
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. Required to induce the systemic acquired resistance (SAR) via the regulation of pathogenesis-related genes expression. Binding to the as-1 element of PR-1 promoter is salicylic acid-inducible and mediated by NPR1. Could also bind to the Hex-motif (5'-TGACGTGG-3') another cis-acting element found in plant histone promoters. {ECO:0000269|PubMed:12897257}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00157 | DAP | Transfer from AT1G22070 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.12659s0060.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | ATHTAGIA | 0.0 | L10209.1 Arabidopsis thaliana transcription factor mRNA, complete cds. | |||
| GenBank | BT026034 | 0.0 | BT026034.1 Arabidopsis thaliana At1g22070 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010459934.1 | 0.0 | PREDICTED: transcription factor TGA3 | ||||
| Swissprot | Q39234 | 0.0 | TGA3_ARATH; Transcription factor TGA3 | ||||
| TrEMBL | Q147Q9 | 0.0 | Q147Q9_ARATH; At1g22070 | ||||
| STRING | Bostr.12659s0060.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3061 | 27 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G22070.1 | 0.0 | TGA1A-related gene 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.12659s0060.1.p |



