 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.19424s0443.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 848aa MW: 97189.6 Da PI: 7.0948 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 81 | 2.1e-25 | 92 | 194 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk............tekerrtraetrtgCkaklkvkkekdgkwe 77
+fY+eY++ +GF++ +++s++sk+++e+++++f Cs++g+++e +k+ e+ +r+ +t+Cka+++vk++ dgkw+
Bostr.19424s0443.1.p 92 SFYQEYSRAIGFNTAIQNSRRSKTTREFIDAKFACSRYGTKREYDKSfnrprarqskqdPENMAGRRTCAKTDCKASMHVKRRPDGKWV 180
5*******************************************99999999877654444444499999******************* PP
FAR1 78 vtkleleHnHelap 91
+++++ eHnHel p
Bostr.19424s0443.1.p 181 IHSFVREHNHELLP 194
***********975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 2.9E-23 | 92 | 194 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 1.8E-24 | 292 | 384 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 9.161 | 572 | 608 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 6.7E-7 | 583 | 610 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 848 aa Download sequence Send to blast |
MDIDLRLHSG DLCKGDDEDR GLDNVLQNEH KLHDADEEDM DIGKIEDVTV EVHTDDSVAM 60 GAPTGELLEY TEGMNLEPLN GMEFESHGEA YSFYQEYSRA IGFNTAIQNS RRSKTTREFI 120 DAKFACSRYG TKREYDKSFN RPRARQSKQD PENMAGRRTC AKTDCKASMH VKRRPDGKWV 180 IHSFVREHNH ELLPAQAVSE QTRKIYAAMA KQFAEYKTVI SLKSDSKSSF EKGRNLSVET 240 GDFKILLDFL SRMQNLNSNF FYAVDLGDDQ RVKNAFWVDG KSRHNYGSFC DVVSLDTTYV 300 RNKYKMPLAI FVGVNQHYQY MVLGCALISD ESAATFSWLM ETWLRAMGGQ APKVLITELD 360 VVMNSIVPEI FPNTRHCLFL WHVLMKVSEN LGQVVKQHDN FMPKFEKCIY KSWKEEDFAR 420 KWWKNLARFG LKEDQWLHSL YEDRKKWAPT YMTDVLLAGM STSQRADSIN AFFDKYMHKK 480 TSVLEFLKLY DTILQDRCEE EAKADSETWN KQPAMKSPSP FEKSVSEVYT PAVFKKFQIE 540 VLGAIACSPR EENRDATCST FRVQDFENNQ DFMVTWKQTK AEVSCICRLF EYKGYLCRHT 600 LNVLQCCHLS SIPSQYILKR WTKDAKSRHF SGEPQQLQTR LLRYNDLCQR ALKLNEEASF 660 SQESYNIAFL AIEGALGNCA GINTSGRSLP DVVTSPTQGL ISVEEDNQNR SAGKTSKKKN 720 PTKKRKVNSE QEVMPVAAPE SLQQMDKLSP RTVGIESYYG TQQSVQGMVQ LNLMAPTRDN 780 FYGNQQTIQG LRQLNSIAPS YDGYYGAQHG IHGQGVDFFR PPNFPYDIRD DPNVRTTQLH 840 EDASRHS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
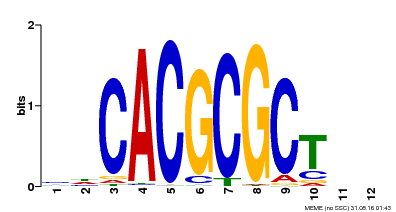
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.19424s0443.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP001306 | 0.0 | AP001306.1 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone:MKA23. | |||
| GenBank | CP002686 | 0.0 | CP002686.1 Arabidopsis thaliana chromosome 3, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020886687.1 | 0.0 | protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X1 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A178VJL5 | 0.0 | A0A178VJL5_ARATH; FHY3 | ||||
| STRING | Bostr.19424s0443.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM118 | 26 | 253 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.19424s0443.1.p |



