 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.20903s0042.3.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 273aa MW: 30564.9 Da PI: 10.3342 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 48.9 | 1.7e-15 | 2 | 40 | 40 | 78 |
EEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 40 qrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
q+ q +rfh+lsefD ekrsCrrrLa hnerrrk+q+
Bostr.20903s0042.3.p 2 QKLITQDTRFHQLSEFDLEKRSCRRRLACHNERRRKPQP 40
56677999****************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51141 | 14.819 | 1 | 38 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:4.10.1100.10 | 6.2E-5 | 2 | 25 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.12E-12 | 5 | 43 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.1E-7 | 7 | 37 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF02466 | 1.4E-10 | 211 | 264 | IPR003397 | Mitochondrial inner membrane translocase subunit Tim17/Tim22/Tim23/peroxisomal protein PMP24 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008361 | Biological Process | regulation of cell size | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0042127 | Biological Process | regulation of cell proliferation | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 273 aa Download sequence Send to blast |
MQKLITQDTR FHQLSEFDLE KRSCRRRLAC HNERRRKPQP TTLFTSHYTR IAPSRYGNAN 60 AAMLKTVLGD PTAWSTARSL MRRSGPCPVK ESHQLMNVFS QESSSFTITC PEMMNNNSTD 120 SSCALSLLSN SNSNPIQQHQ QQLQTKTNIW RPPSGFDSMI VDRVTMAQPL PISSHHQYLN 180 QTLEFMPGEN SNSHYMSPKD VPRVERNVAL PGLIKTLKMM GSHGLTFAAI GGVFIGVEQL 240 VQNFRAKRDV FNGAIGGFVA GASVLGYKGW WF* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00412 | DAP | Transfer from AT3G57920 | Download |
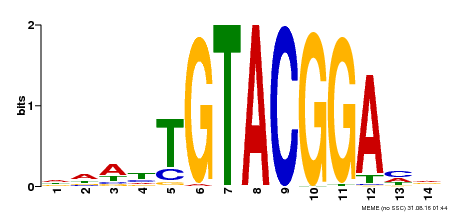
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.20903s0042.3.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR157. {ECO:0000305|PubMed:12202040}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006291402.1 | 1e-102 | squamosa promoter-binding-like protein 15 | ||||
| Swissprot | Q9M2Q6 | 1e-94 | SPL15_ARATH; Squamosa promoter-binding-like protein 15 | ||||
| TrEMBL | R0HGL0 | 1e-101 | R0HGL0_9BRAS; Uncharacterized protein | ||||
| STRING | Bostr.20903s0042.1.p | 0.0 | (Boechera stricta) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G57920.1 | 1e-82 | squamosa promoter binding protein-like 15 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.20903s0042.3.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



