 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.22252s0001.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 214aa MW: 24544.6 Da PI: 5.7524 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.3 | 2.6e-16 | 94 | 138 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+W+teE+ l++d+ +++G+g+Wk+I+r+ k+R++ q+ s+ qky
Bostr.22252s0001.1.p 94 PWSTEEHRLFLDGLEKYGKGDWKSISRECVKTRSPTQVASHAQKY 138
7*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 1.3 | 3 | 54 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.22E-6 | 6 | 60 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.135 | 86 | 143 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 6.68E-17 | 90 | 143 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.9E-10 | 91 | 141 | IPR001005 | SANT/Myb domain |
| TIGRFAMs | TIGR01557 | 5.7E-16 | 91 | 142 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-10 | 92 | 138 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.2E-12 | 94 | 138 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.39E-11 | 94 | 139 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 214 aa Download sequence Send to blast |
MASIPQWTRD DDKRFELALV QFPEGSPSFL ENIAQLLQKP LEEVKYYYEA LVDDVALIES 60 GKVALPKYPE DDDVSLKEAT QSKKLRKKIR KGIPWSTEEH RLFLDGLEKY GKGDWKSISR 120 ECVKTRSPTQ VASHAQKYFL RQELGIKRGK RLSIHDMTLP GDAENVTVPG SNLNSKGQQL 180 QFGDQIPPDQ YYHYSQDNFG LFDDDDDKWF SKL* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 82 | 90 | KKLRKKIRK |
| 2 | 83 | 89 | KLRKKIR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that coordinates abscisic acid (ABA) biosynthesis and signaling-related genes via binding to the specific promoter motif 5'-(A/T)AACCAT-3'. Represses ABA-mediated salt (e.g. NaCl and KCl) stress tolerance. Regulates leaf shape and promotes vegetative growth. {ECO:0000269|PubMed:26243618}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00344 | DAP | Transfer from AT3G10580 | Download |
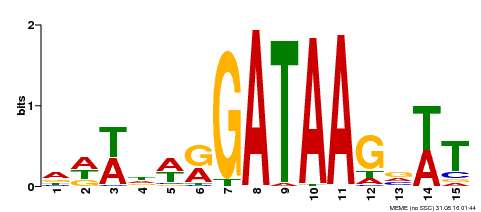
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.22252s0001.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salicylic acid (SA) and gibberellic acid (GA) (PubMed:16463103). Triggered by dehydration and salt stress (PubMed:26243618). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:26243618}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC011560 | 7e-77 | AC011560.6 Arabidopsis thaliana chromosome 3 BAC F13M14 genomic sequence, complete sequence. | |||
| GenBank | ATAC013428 | 7e-77 | AC013428.6 Arabidopsis thaliana chromosome III BAC F18K10 genomic sequence, complete sequence. | |||
| GenBank | CP002686 | 7e-77 | CP002686.1 Arabidopsis thaliana chromosome 3, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010486370.1 | 1e-103 | PREDICTED: transcription factor DIVARICATA-like | ||||
| Swissprot | Q9FNN6 | 4e-39 | SRM1_ARATH; Transcription factor SRM1 | ||||
| TrEMBL | D7L911 | 7e-98 | D7L911_ARALL; Uncharacterized protein | ||||
| STRING | Bostr.22252s0001.1.p | 1e-156 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM14731 | 7 | 19 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G10580.2 | 1e-100 | MYB_related family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.22252s0001.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



