 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.25993s0059.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 933aa MW: 103997 Da PI: 5.4287 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 135.1 | 2.2e-42 | 140 | 217 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+Ceadls++k+yhrrhkvCe+hska++++v g++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
Bostr.25993s0059.2.p 140 VCQVENCEADLSKVKDYHRRHKVCEMHSKATSAMVGGIMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTNP 217
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 2.4E-34 | 134 | 202 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.633 | 138 | 215 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.7E-39 | 139 | 219 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.8E-31 | 141 | 214 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 2.0E-7 | 694 | 822 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.82E-7 | 723 | 824 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 7.75E-9 | 725 | 821 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 933 aa Download sequence Send to blast |
MEGEAQHFYG FRAVDLRSVG KRSVEWDLND WKWDGDLFVA TQLNPGGSET TGRQFFPIGN 60 SSNSSSSCSD EGNNSDLMVR STREVEKKRR VVTIEGDNNG GDGGLTLKLG DNGCDLNGER 120 EGNYAAKKTK FGGGVTNRAV CQVENCEADL SKVKDYHRRH KVCEMHSKAT SAMVGGIMQR 180 FCQQCSRFHV LQEFDEGKRS CRRRLAGHNK RRRKTNPEPG ANGNPSDDQS SNYLLISLLK 240 ILSNMHTTRS DPTGDQDLMS HLLKSLVSHA GEQLGKNLVE LLLQGGSRGS LNLGNSGLLA 300 IEEAPQEDLK QLSVNVPEIP QQELYANGTA TENRSEKQVK MNDFDLNDIY IDSDNTDIER 360 SPPPTNPATS SLDYPSWIHQ SSPPQTSRNS DSASDQSPSS SSEDAQMRTG RIVFKLFGKE 420 PNDFPVVLRG QILDWLSHSP TDMESYIRPG CIVLTIYLRQ AETAWEELSD DLGFSLRKLL 480 DLSDDPLWTT GWIYVRVQNQ LAFVYNGQVV VDTSLPLKSR DYSHIISVKP LAVAATGKAQ 540 FTVKGINIRR RGTSLLCAVE GKYLIQETTH DSTTEENDDL KENNEIVECV NFSCDMPIIS 600 GRGFMEIEDQ GLSSSFFPFL VVEEDDVCSE IRILETTLEF TGTDSAKQAM DFIHEIGWLL 660 HRSKLGESDP NPDVFPLIRF QWLIEFSMDR EWCAVIRKLL NMFFDGAVGD FSSSSNATLS 720 ELCLLHRAVR KNSKPLVETL LRYVPKQQRN SLFRPDAAGP AGLTPLHIAA GKDGSEDVLD 780 ALTEDPAMVG IEAWKTSRDS TGFTPEDYAR LRGHFSYIHL IQRKINKKST TEDHVVVNIP 840 VSFSDREQKE PKSGSVTSVL EITQINQLPC KLCDHKLVYG TTRRSVAYRP AMLSMVAIAA 900 VCVCVALLFK SCPEVLYVFQ PFRWELLDYG TS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 5e-32 | 134 | 214 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
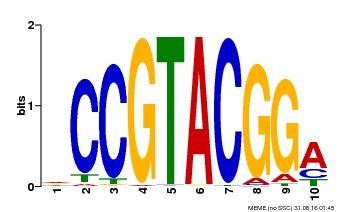
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00603 | PBM | Transfer from AT2G47070 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.25993s0059.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC815948 | 0.0 | KC815948.1 Arabis alpina SQUAMOSA promoter binding protein-like protein 1 (SPL1) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023639628.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Refseq | XP_023639629.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | D7LFY4 | 0.0 | D7LFY4_ARALL; Uncharacterized protein | ||||
| STRING | Bostr.25993s0059.1.p | 0.0 | (Boechera stricta) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.25993s0059.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



