 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.26326s0173.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 272aa MW: 30483.3 Da PI: 9.0899 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 93.8 | 1.2e-29 | 145 | 204 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
++Dgy+WrKYGqK ++++ pr+Y+rC+++ C vkkkv+rsaedp++v+ tYeg+Hnh+
Bostr.26326s0173.1.p 145 VKDGYQWRKYGQKITRDNPSPRAYFRCSFSpACLVKKKVQRSAEDPSFVVATYEGTHNHP 204
58***************************99****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.1E-27 | 138 | 206 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.32E-25 | 142 | 206 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.6E-33 | 145 | 205 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 8.0E-24 | 146 | 204 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 26.923 | 147 | 206 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0002237 | Biological Process | response to molecule of bacterial origin | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0031347 | Biological Process | regulation of defense response | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 272 aa Download sequence Send to blast |
MDYDPNTNPF DLHFSGKLPK REVSSSGSAD VKRKCLVKDE KTYMLQEELN RVNSENKKLT 60 ETLARVCEKY YALHNLLEEL QSRKSPENVN FQNEQLTRKR KQDLDDFVSS PIGLSCGTIE 120 NITSGKATVS TAYFHADKSD TSLTVKDGYQ WRKYGQKITR DNPSPRAYFR CSFSPACLVK 180 KKVQRSAEDP SFVVATYEGT HNHPGPHASG SRTVKLDLVQ SGLEPVEEKK ERGTIQEVLV 240 QHMASSLTKD PKFTAALAAA LSGRFIEQSR T* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 4e-20 | 147 | 203 | 19 | 74 | Probable WRKY transcription factor 4 |
| 2lex_A | 4e-20 | 147 | 203 | 19 | 74 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 96 | 101 | TRKRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00068 | PBM | Transfer from AT2G25000 | Download |
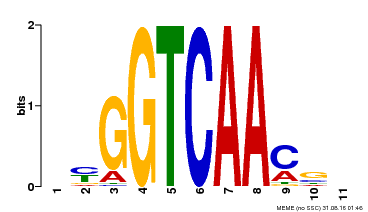
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.26326s0173.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK227987 | 0.0 | AK227987.1 Arabidopsis thaliana mRNA for putative WRKY-type DNA binding protein, complete cds, clone: RAFL14-52-F05. | |||
| GenBank | BT004611 | 0.0 | BT004611.1 Arabidopsis thaliana At2g25000 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_180072.1 | 1e-176 | WRKY DNA-binding protein 60 | ||||
| Refseq | XP_010472507.1 | 1e-176 | PREDICTED: probable WRKY transcription factor 60 | ||||
| Swissprot | Q9SK33 | 1e-177 | WRK60_ARATH; Probable WRKY transcription factor 60 | ||||
| TrEMBL | Q0WSE5 | 1e-174 | Q0WSE5_ARATH; Putative WRKY-type DNA binding protein | ||||
| STRING | Bostr.26326s0173.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6584 | 19 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G25000.1 | 1e-179 | WRKY DNA-binding protein 60 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.26326s0173.1.p |



