 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.28243s0053.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 461aa MW: 52378.4 Da PI: 5.7284 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.5 | 1e-12 | 294 | 339 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er RR+++N++f Lr ++P+ K++K++ Le Av+YI++L
Bostr.28243s0053.1.p 294 NHVEAERMRREKLNHRFYALRAVVPNV-----SKMDKMSLLEDAVRYINEL 339
799***********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 2.4E-51 | 33 | 226 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.552 | 290 | 339 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 6.15E-17 | 293 | 360 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.19E-13 | 293 | 340 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 1.2E-15 | 294 | 360 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 4.6E-10 | 294 | 339 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.8E-14 | 296 | 345 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005509 | Molecular Function | calcium ion binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 461 aa Download sequence Send to blast |
MNTDGNLYMM ESLLTSDLWQ PLPPANPSLE TTLQKRLHAV LNGTHEPWTY AIFWKPSYYD 60 FSGESMLKWG DGVYSGRDKE EKTRRRKRKM MIPSSPEEKE RRNKVLRELN FMISGEAFPV 120 VEDDDVHDDD DDVEVTDMEW FFLVSMTWSF GNGSGLPGKA FTTYNPVWVT GSDQIYESGC 180 DRAKQGGGLG LQTIVCIPCD NGVLELGSME QIRQNSDIFN KTRFLFNFQG SKDFSGALTT 240 SIQFHSSTDN PNPDPSQVYQ PIQTNRIDAT VIPKKKPGKK RGRKPAHGRE EPLNHVEAER 300 MRREKLNHRF YALRAVVPNV SKMDKMSLLE DAVRYINELK SKAENAESEK NAIQIQLDEL 360 NEIMGRQNAI SSVCNDKENV SELKIEVKIM GNDVMIRVES GKRKHPGARL MNALMDLELE 420 VDHAAISVMN DLMIQQATVT MGLRIYEQEQ LRDTLISKIS * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 9e-49 | 30 | 228 | 6 | 194 | Transcription factor MYC3 |
| 4rqw_B | 9e-49 | 30 | 228 | 6 | 194 | Transcription factor MYC3 |
| 4rs9_A | 9e-49 | 30 | 228 | 6 | 194 | Transcription factor MYC3 |
| 4yz6_A | 9e-49 | 30 | 228 | 6 | 194 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00547 | DAP | Transfer from AT5G46830 | Download |
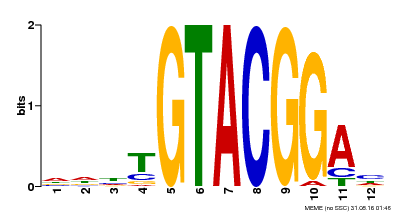
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.28243s0053.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010441529.1 | 0.0 | PREDICTED: transcription factor bHLH28-like | ||||
| Swissprot | Q9LUK7 | 0.0 | BH028_ARATH; Transcription factor bHLH28 | ||||
| TrEMBL | D7MQN9 | 0.0 | D7MQN9_ARALL; Predicted protein | ||||
| STRING | Bostr.28243s0053.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13953 | 16 | 24 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G46830.1 | 0.0 | NACL-inducible gene 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.28243s0053.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



