 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.30275s0060.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 924aa MW: 104635 Da PI: 7.1427 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 162 | 1.1e-50 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahsee 90
+e +rwl+++ei+a+L n++ +++ ++ + pksg+++L++rk++r+frkDG+swkkkkdgkt++E+he+LKvg+ e +++yYah+++
Bostr.30275s0060.1.p 30 DEaYTRWLRPNEIHALLCNHHFFTINVKPVNLPKSGTIVLFDRKMLRNFRKDGHSWKKKKDGKTIKEAHEHLKVGNEERIHVYYAHGDD 118
45589************************************************************************************ PP
CG-1 91 nptfqrrcywlLeeelekivlvhylevk 118
nptf rrcywlL++++e+ivlvhy+e++
Bostr.30275s0060.1.p 119 NPTFVRRCYWLLDKSKEHIVLVHYRETH 146
*************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 76.134 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 4.2E-72 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.1E-44 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.68E-13 | 374 | 460 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 5.48E-14 | 560 | 670 | No hit | No description |
| Pfam | PF12796 | 3.1E-7 | 560 | 640 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.7E-15 | 561 | 673 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.71E-15 | 569 | 681 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 13.708 | 578 | 643 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 2.7E-6 | 611 | 640 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.808 | 611 | 643 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 6.595 | 760 | 786 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 260 | 775 | 797 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 4.7E-4 | 798 | 820 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.651 | 799 | 823 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 6.4E-4 | 801 | 820 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 6.8 | 875 | 897 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.114 | 876 | 905 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.22 | 877 | 897 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 924 aa Download sequence Send to blast |
MSGVGSGKLI GSEIHGFHTL QDLDIQTMLD EAYTRWLRPN EIHALLCNHH FFTINVKPVN 60 LPKSGTIVLF DRKMLRNFRK DGHSWKKKKD GKTIKEAHEH LKVGNEERIH VYYAHGDDNP 120 TFVRRCYWLL DKSKEHIVLV HYRETHEVQA APATPGNSYS SSISDHLSPK LVAEDINSDV 180 RNTCNTGFDV RSNSLGARNH EIRLHEINTL DWDELLVPAD ISNQSHPTEE DMLYLTGQLQ 240 TAPRGSAKQG NHLAGYNGSV DIPSFPGLED PVYQNNNSCG AGEFSSQHMH CGVEPNLQRR 300 DSSATVADQP GDALLNNGYG SQDSFGRWVN NFISDSPGSV DDPSLEAVCT PGQESSTTPA 360 GLHSHSNIPE QVFNITDVSP AWAYSTEKTK ILVTGFFHDS FQHFGISNLF CICGELRVPA 420 EFLQMGVYRC FLPPQSPGVV NLYLSVDGNK PISQLFSFEH RSVPVIETAI PQDDQLYMWE 480 EFEFQVRLAH LLFTSSNKIS ILTSKISPDN LQEAKKLASR TSHLLNSWAY LMKSIQANEV 540 PFDQARDHLF ELTLKNRLKE WLLEKVIENR NTKEYDSKGL GVIHLCAILG YTWSILLFSW 600 ANISLDFRDK HGWTALHWAA YYGREKMVAA LLSAGARPNL VTDPTKEFLG GCTAADLAQQ 660 KGYEGLAAFL AEKCLVAQFK DMQMAGNISG NLETIKAEKS SNPGNANEEE QSLKDTLAAY 720 RTAAEAAARI QGAFREHELK VRSSAVRFAS KEEEAKNIVA AMKIQHAFRN FETRRKIAAA 780 ARIQYRFQTW KMRREFLNMR NKAIRIQAAF RGFQVRRQYQ KITWSVGVLE KAILRWRLKR 840 KGFRGLQVSQ PEEKEGSDEE VEEFYKTSQK QAEERLERSV VKVQAMFRSK KAQQDYRRIK 900 LAHEEAELEY DGMQEHDQMA MES* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
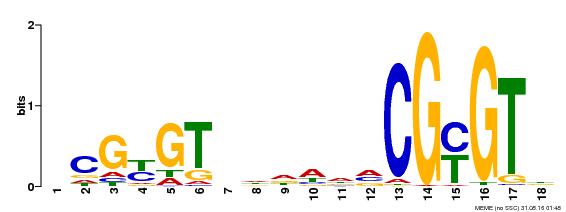
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.30275s0060.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY128295 | 0.0 | AY128295.1 Arabidopsis thaliana AT4g16150/dl4115w mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006282550.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X1 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | R0GWD2 | 0.0 | R0GWD2_9BRAS; Uncharacterized protein | ||||
| STRING | Bostr.30275s0060.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4021 | 27 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.30275s0060.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



