 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.7128s0689.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1043aa MW: 115499 Da PI: 8.0116 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.4 | 1.3e-40 | 132 | 209 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ak+yhrrhkvCevhska+++lv +l+qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+++
Bostr.7128s0689.1.p 132 MCQVDNCTEDLSHAKDYHRRHKVCEVHSKATKALVGKLMQRFCQQCSRFHLLSEFDEGKRSCRRRLAGHNRRRRKTTQ 209
6**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.9E-34 | 125 | 194 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.214 | 130 | 207 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.22E-37 | 131 | 209 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.1E-28 | 133 | 206 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 2.98E-9 | 794 | 944 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 3.76E-9 | 794 | 937 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.9E-7 | 833 | 940 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 9.596 | 837 | 950 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1043 aa Download sequence Send to blast |
MDEVGAQVAT PMFIHPSLYP MGRKRDLYYP MSNRLVPSQP QPQPQRRDEW NSKMWDWDSR 60 RFEAKPVDAQ TLRLGNETQQ FDLTSRNRSG EERGLDLNLG SGLTAVEDTT TTQNIRPNKK 120 VRSGSPGGNY PMCQVDNCTE DLSHAKDYHR RHKVCEVHSK ATKALVGKLM QRFCQQCSRF 180 HLLSEFDEGK RSCRRRLAGH NRRRRKTTQQ EEIASGVVVP GNRDNTSNAN MDLMALLTAL 240 ACAQGRNEVK PASSPAVPDR EQLLQILDKI NALPLPMDLV SKLNNIGSLA RKNLDQPKAN 300 PPNDMNGASP STMDLLAVLS STLGSSSPDA LAILSQGGFG NKDSDKTKLS SYDHGVTTNV 360 EKRTFGFSSG GGERSSSSNQ SPSQDSDSRA QDTRSSLSLQ LFTSSPEDES RPTVASSRKY 420 YSSASSNPVE DRSPSSSPVM QELFPLQTSP ETMRSKNHNN SSPRTGCLPL ELFGASNRGT 480 ANPNFKGFGQ QSGYASSGSD YSPPSLNSDA QDRTGKIVFK LLDKDPSQLP GTLRSEIYNW 540 LSNIPSEMES YIRPGCVVLS VYIAMSPAAW EQLEQNLLQR LGVLLQNSQS DFWRNARFIV 600 NTGRQLASHK NGRVRCSKSW RTWNSPELIS VSPVAVVAGE ETSLIVRGRS LTNDGISIRC 660 THMGSYMSME VTGAACRQAV FDELNVNSFK VQNAQPGFLG RCFIEVENGF RGDSFPLIIA 720 NASICKELNR LEEEFHRKSE DTTEEQAQSS DCRPTSREEV VCFLNELGWL FQKNQTSELR 780 EQPDFSLARF KFLLVCSVER DYCALIRTLL DMLVERNLVN DELNAEALDM LAEIQLLNRA 840 VKRKSTKMVE QLIHYSVNPS ALDSSNNFVF LPNITGPGGI TPLHLAACTS GSDDMVDLLT 900 NDPQEIGLSN WNSLCDATGK TPYSYAAMRN NHSYNSLVAR KLADKRNKQV SLNIENEVVD 960 QNGLSKRLSS EMNKSSCASC ATVALKYQRR VSGSHRLFPT PIIHSMLAVA TVCVCVCVFM 1020 HAFPIVRQGS HFSWGGLDYG SI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-32 | 123 | 206 | 3 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
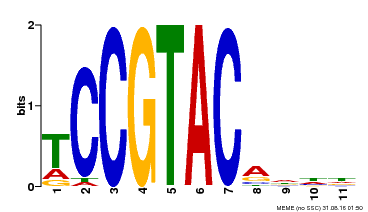
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.7128s0689.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK226907 | 0.0 | AK226907.1 Arabidopsis thaliana mRNA for squamosa promoter binding protein-like 14, clone: RAFL09-16-H09. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006303847.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | R0IEQ3 | 0.0 | R0IEQ3_9BRAS; Uncharacterized protein | ||||
| STRING | Bostr.7128s0689.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.7128s0689.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



