 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bostr.8819s0047.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Boechereae; Boechera
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 683aa MW: 76363.3 Da PI: 5.6184 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 160.4 | 1.2e-49 | 78 | 227 | 1 | 145 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasasp 90
gg+g+++++kE+E++k+RER+RRai++++++GLR++Gn++lp+raD+n+V++AL+reAGw v++DGttyr++ +p+ ++++ ++s+
Bostr.8819s0047.1.p 78 GGKGKREREKEKERTKLRERHRRAITSRMLSGLRQYGNFPLPARADMNDVIAALAREAGWSVDADGTTYRQSHQPN----HVVQYPTRSI 163
6899************************************************************************....8889999999 PP
DUF822 91 esslq.sslkssalaspvesysaspksssfpspssldsislasa........asllpvlsvlsl 145
es+l+ s+ k++a+a+ +++++++ ++ +sp+slds+ +a++ + +p++sv +l
Bostr.8819s0047.1.p 164 ESPLSsSTFKNCAKAALDCQQHSVLRIDENLSPVSLDSVVIAESdhpgngryTGASPITSVGCL 227
999988899********************************99888888777677777777776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.1E-47 | 79 | 226 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 4.59E-159 | 246 | 680 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 1.6E-166 | 249 | 679 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 2.3E-81 | 254 | 643 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.7E-53 | 285 | 299 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.7E-53 | 306 | 324 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.7E-53 | 328 | 349 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.7E-53 | 421 | 443 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.7E-53 | 494 | 513 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.7E-53 | 528 | 544 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.7E-53 | 545 | 556 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.7E-53 | 563 | 586 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 3.7E-53 | 601 | 623 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 683 aa Download sequence Send to blast |
MHTLNNTTGS QDPNLDPIPD PDQFPNRNHN HPQSRRPRGF AAAAAAASIG PTESDVNNGN 60 IAGIGGGEGS GGGGGGGGGK GKREREKEKE RTKLRERHRR AITSRMLSGL RQYGNFPLPA 120 RADMNDVIAA LAREAGWSVD ADGTTYRQSH QPNHVVQYPT RSIESPLSSS TFKNCAKAAL 180 DCQQHSVLRI DENLSPVSLD SVVIAESDHP GNGRYTGASP ITSVGCLEAN QLIQDVHSTE 240 PRNDFTESFY VPVYAMLPVG NINNFGQLVD PEGVRQELSY MKSLNVDGVV IDCWWGIVEG 300 WNPQKYVWSG YRELFNLIRD FKLKLQVVMA FHEYGGNASG NVIISLPQWV LEIGKDNPDI 360 FFTDREGRRS FECLNWSIDK ERVLHGRTGI EVYFDFMRSF RSEFDDLFVE GLIAAVEIGL 420 GASGELKYPS FPERMGWIYP GIGEFQCYDK YSQSNLQKEA ESRGFVFWGK GPENAGQYNS 480 QPHETGFFQE RGEYDSYYGR FFLNWYSQLL IGHAENVLSL ANLALEETKI IVKIPAIYWS 540 YKTASHAAEL TAGYYNPSNR DGYSLLFETL KKYSVTVKFV CPGPQMSPNE HEEALADPEG 600 LSWQVINAAW DKRLLIGGEN AITCFDREGC MRLIDIAKPR NHPDNYHFSF FTYRQPSPLV 660 QGSTCFPDLD DFIKSMHGDI QR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-116 | 250 | 679 | 11 | 444 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
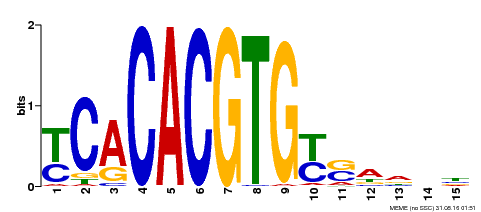
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bostr.8819s0047.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK117140 | 0.0 | AK117140.1 Arabidopsis thaliana At5g45300 mRNA for putative beta-amylase, complete cds, clone: RAFL16-68-D16. | |||
| GenBank | BT006482 | 0.0 | BT006482.1 Arabidopsis thaliana At5g45300 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010494622.1 | 0.0 | PREDICTED: beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | D7MUR1 | 0.0 | D7MUR1_ARALL; Beta-amylase | ||||
| STRING | Bostr.8819s0047.1.p | 0.0 | (Boechera stricta) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10888 | 28 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bostr.8819s0047.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



