 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi1g14750.1.p | ||||||||
| Common Name | BRADI_1g14750, LOC100831213 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 222aa MW: 23360.4 Da PI: 7.7682 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 14.6 | 9.5e-05 | 80 | 102 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
y+C+ Cgk F++ L H +H
Bradi1g14750.1.p 80 YTCSVCGKAFPSYQALGGHKASH 102
9***********99999998887 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF13912 | 3.6E-13 | 80 | 104 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 10.783 | 80 | 107 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.3E-10 | 80 | 102 | No hit | No description |
| SMART | SM00355 | 0.007 | 80 | 102 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 7.6E-4 | 80 | 102 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 82 | 102 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 2.3E-10 | 137 | 164 | No hit | No description |
| Pfam | PF13912 | 3.8E-14 | 141 | 166 | IPR007087 | Zinc finger, C2H2 |
| PROSITE profile | PS50157 | 9.349 | 142 | 164 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.12 | 142 | 164 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 144 | 164 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006979 | Biological Process | response to oxidative stress | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009644 | Biological Process | response to high light intensity | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010117 | Biological Process | photoprotection | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0015979 | Biological Process | photosynthesis | ||||
| GO:0035264 | Biological Process | multicellular organism growth | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 222 aa Download sequence Send to blast |
MAVEAVLDAA AIPWPAASDQ QEETTTQHGG LQYCAEGWGK RKRTRRQQQQ PPTEEEHLAL 60 SLLMLARGHR DGTSSLQAQY TCSVCGKAFP SYQALGGHKA SHRPKASPPF IGAVDEPAAN 120 NTPSPAASSS TCSGAATAGK VHECSVCKKT FPTGQALGGH KRCHYEGPLG GSGSASRGFD 180 LNLPALPDII VAEPRYCVPA AEEEEVLSPM AFKKPRLMIP A* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Potential transcription factor which binds the nonamer motif 5'-CATCCAACG-3'. Possibly involved in regulating transcription of the histone H3 and H4 genes. {ECO:0000269|PubMed:8223628}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00162 | DAP | Transfer from AT1G27730 | Download |
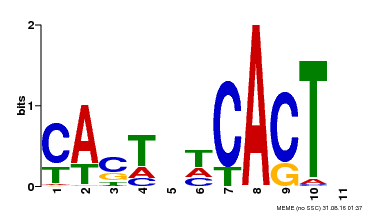
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi1g14750.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003562286.1 | 1e-163 | zinc finger protein 1 | ||||
| Swissprot | Q42430 | 2e-40 | ZFP1_WHEAT; Zinc finger protein 1 | ||||
| TrEMBL | I1GQB7 | 1e-161 | I1GQB7_BRADI; Uncharacterized protein | ||||
| STRING | BRADI1G14750.1 | 1e-162 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1022 | 36 | 113 | Representative plant | OGRP131 | 15 | 149 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G27730.1 | 3e-36 | salt tolerance zinc finger | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi1g14750.1.p |
| Entrez Gene | 100831213 |



