 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi1g23340.2.p | ||||||||
| Common Name | BRADI_1g23340 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 530aa MW: 57577.8 Da PI: 6.5851 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102.2 | 3e-32 | 151 | 208 | 2 | 60 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS-- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhek 60
+DgynWrKYGqK+vkgse+prsYY+Ct+++C+vkk +er+a d++++e++Y+g+Hnh+k
Bradi1g23340.2.p 151 EDGYNWRKYGQKHVKGSENPRSYYKCTHPNCEVKKLLERAA-DGQITEVVYKGRHNHPK 208
8****************************************.***************85 PP
| |||||||
| 2 | WRKY | 107.6 | 6.3e-34 | 324 | 382 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
ldDgy+WrKYGqK+vkg+++prsYY+Cts+gCpv+k+ver+++dpk v++tYeg+Hnhe
Bradi1g23340.2.p 324 LDDGYRWRKYGQKVVKGNPNPRSYYKCTSTGCPVRKHVERASHDPKSVITTYEGKHNHE 382
59********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 6.3E-28 | 143 | 209 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 24.122 | 145 | 209 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.81E-25 | 148 | 209 | IPR003657 | WRKY domain |
| SMART | SM00774 | 4.4E-35 | 150 | 208 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 9.9E-25 | 151 | 207 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 5.1E-38 | 309 | 384 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.81E-30 | 316 | 384 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 38.871 | 319 | 384 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.4E-39 | 324 | 383 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.7E-26 | 325 | 382 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009961 | Biological Process | response to 1-aminocyclopropane-1-carboxylic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 530 aa Download sequence Send to blast |
MKVEPSPTTG SLGMAAIMHK SAHPDILPSP RDKSVLSAHE DGGSRDFEFK PHLNSSSQSM 60 APAMSDLKNH EHSMQNQSTN PSSSSNMVIE YRPPCSREST LAVNVSSAQD QLGLTDSMPV 120 DVGTSELHQM NNSENAMQEP QSEHATEKSA EDGYNWRKYG QKHVKGSENP RSYYKCTHPN 180 CEVKKLLERA ADGQITEVVY KGRHNHPKPQ PNRRLAGGAV PSNQGEDRND GLAAIDDKSS 240 NVLSILGNPV HSTGMAEPVP GSASDDDIDA GAGRPYPGDD ATEDDDLESK RRKMESAGID 300 AALMGKPNRE PRVVVQTVSE VDILDDGYRW RKYGQKVVKG NPNPRSYYKC TSTGCPVRKH 360 VERASHDPKS VITTYEGKHN HEVPAARNAS HEMPAPPMKN AVHPINSNMP SSIGGMMRAC 420 EARNFTNQYS QASETETISL DLGVGISPNH REATNQIQSS VPDQMQYQMQ PMASVYGNMR 480 LPAMAMPTVQ GHAAGSIYGS REEKGNEGFT FKATPMDHSA NLCYSGAGI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 3e-37 | 151 | 385 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 3e-37 | 151 | 385 | 8 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in starch synthesis (PubMed:12953112). Acts as a transcriptional activator in sugar signaling (PubMed:16167901). Interacts specifically with the SURE and W-box elements, but not with the SP8a element (PubMed:12953112). {ECO:0000269|PubMed:12953112, ECO:0000269|PubMed:16167901}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00455 | DAP | Transfer from AT4G26640 | Download |
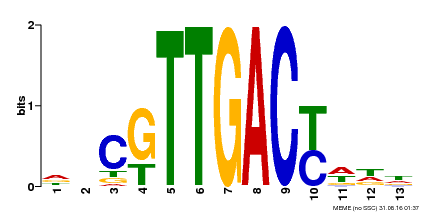
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi1g23340.2.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by sugar. {ECO:0000269|PubMed:12953112}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM201384 | 0.0 | KM201384.1 Stipa purpurea WRKY17 transcription factor mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024315261.1 | 0.0 | WRKY transcription factor SUSIBA2 isoform X2 | ||||
| Swissprot | Q6VWJ6 | 0.0 | WRK46_HORVU; WRKY transcription factor SUSIBA2 | ||||
| TrEMBL | I1GSZ0 | 0.0 | I1GSZ0_BRADI; Uncharacterized protein | ||||
| STRING | BRADI1G23340.1 | 0.0 | (Brachypodium distachyon) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G26640.1 | 1e-104 | WRKY family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi1g23340.2.p |



