 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi1g27170.1.p | ||||||||
| Common Name | BRADI_1g27170, LOC100841919 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 909aa MW: 102350 Da PI: 9.1942 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 163 | 5.5e-51 | 34 | 146 | 5 | 117 |
CG-1 5 kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrr 97
+rw +++ei+a+L+n++++++++++ + p sg+++Ly+rk+vr+frkDG++wkkkkdgktv+E+hekLK+g+ e +++yYa++e+np+f rr
Bradi1g27170.1.p 34 PARWFRPNEIYAVLANHARFKVHAQPIDMPVSGTIVLYDRKVVRNFRKDGHNWKKKKDGKTVQEAHEKLKIGNEERVHVYYARGEDNPNFFRR 126
68******************************************************************************************* PP
CG-1 98 cywlLeeelekivlvhylev 117
cywlL++e e+ivlvhy+++
Bradi1g27170.1.p 127 CYWLLDKEAERIVLVHYRQT 146
*****************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 72.692 | 26 | 152 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.4E-60 | 29 | 147 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.8E-44 | 33 | 145 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 8.82E-11 | 358 | 445 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 1.52E-17 | 541 | 651 | No hit | No description |
| SuperFamily | SSF48403 | 8.24E-19 | 550 | 657 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 2.9E-19 | 553 | 656 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 20.447 | 559 | 663 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 8.5E-8 | 565 | 655 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 6.4E-6 | 592 | 621 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.781 | 592 | 624 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 150 | 631 | 660 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 2.34E-8 | 743 | 808 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 3 | 757 | 779 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.455 | 759 | 784 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0065 | 760 | 778 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0095 | 780 | 802 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.493 | 781 | 805 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.005 | 782 | 802 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.41 | 861 | 883 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.608 | 863 | 891 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045944 | Biological Process | positive regulation of transcription from RNA polymerase II promoter | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0001077 | Molecular Function | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 909 aa Download sequence Send to blast |
MAGAAGRDPL LRSEIHGFIT YADLNFEKLK AEAPARWFRP NEIYAVLANH ARFKVHAQPI 60 DMPVSGTIVL YDRKVVRNFR KDGHNWKKKK DGKTVQEAHE KLKIGNEERV HVYYARGEDN 120 PNFFRRCYWL LDKEAERIVL VHYRQTSEEN AIAHPSTEEA AEVPTMNRSQ YYASPPTSAD 180 SASVHTELSF SPPVPEEINS HGGSAISNGT DGSTLEEFWV HLLESSMKND TSSSGGSMAF 240 SQQIKYRPKD SENNSNTTSN AVLVSPPNVM PEAYPTNHVP ANHVGALKHQ GDQLQYLVTL 300 DVDSQSERFV NTLERTPVDS NIPSDVPARE NSLGLWKYLD DDSPCLGDNI VSNERLFNIT 360 DFSPEWALST EHTKILVVGY YYEQHKHLAG SSMYGVFGDN CVAADMIQSG VYRFMAGPHT 420 PGRVDFYLTL DGKTPISEVL SFEYRSMPGD SLKSDLKPLE DENKKSKLQM QMRLARLMFA 480 TNKKKIAPKL LVEGTRVSNL ISASPEKEWV DLWKIASDSE GTCVPATEDL LELVLRNRLQ 540 EWLLERVIGG HKSTGRDDLG QGPIHLCSFL GYTWAIRLFS SSGFSLDFRD SSGWTALHWA 600 AYHGRERMVA ALLSAGANPS LVTDPTAMSP AGCTPADLAA KQGYVGLAAY LAEKGLTAHF 660 ESMSLTKDTK RSPSRTKLTK VQSDKFENLT EQELCLKESL AAYRNAADAA SNIQAALRDR 720 TLKLQTKAIL ANPELQAAEI VAAMRIQHAF RNYNRKKVMR AAAQIQNHFR TWKVRKNFTN 780 MRRQAIRIQA AYRGHQVRRQ YRKVIWSVGV VEKAILRWRK KRKGLRGIGN GMPVEMTVDV 840 EPASTAEEDY FQASRQQAED RFNRSVVRVQ ALFRCHRAQH EYRRMRIAHE EARLEFSGEP 900 QQGPARRS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
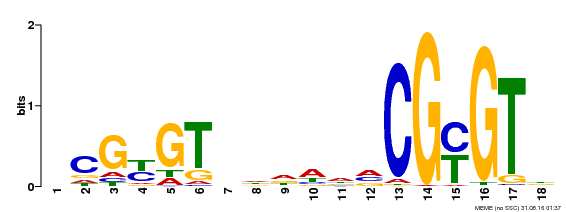
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi1g27170.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK375447 | 0.0 | AK375447.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3094H17. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003563086.1 | 0.0 | calmodulin-binding transcription activator CBT | ||||
| Swissprot | Q7XHR2 | 0.0 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | I1GUA4 | 0.0 | I1GUA4_BRADI; Uncharacterized protein | ||||
| STRING | BRADI1G27170.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9831 | 29 | 33 | Representative plant | OGRP7351 | 11 | 16 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi1g27170.1.p |
| Entrez Gene | 100841919 |



