 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi2g08080.1.p | ||||||||
| Common Name | BRADI_2g08080, LOC100844558 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 618aa MW: 67416.2 Da PI: 7.0707 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.6 | 9.7e-13 | 463 | 509 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
Bradi2g08080.1.p 463 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAIAYITDLQ 509
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 6.9E-49 | 60 | 245 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.806 | 459 | 508 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.17E-15 | 462 | 513 | No hit | No description |
| SuperFamily | SSF47459 | 2.22E-18 | 462 | 524 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.2E-10 | 463 | 509 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 5.4E-18 | 463 | 521 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-16 | 465 | 514 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 618 aa Download sequence Send to blast |
MVMKMEVEED GANGGTGGTW TEEDRALSTT VLGRDAFAYL TKGGGAISEG LVAASLPADL 60 QNKLQELIES EHPHGGWNYA IFWQLSRTKS GDLVLGWGDG SCREPHDGEV GGAASVGNDD 120 ANQRMRKRVL QRLHTAFGGA DEEDYAPGID QVTDTEMFFL ASMYFAFPRR AGGPGQVFAA 180 GMPLWIPNTD RNVFPVNYCY RGYLASTAGF RTIVLVPFET GVLELGSMQQ VVESPDALQA 240 IKAVFAGSGN IVQRREGNGH IEKSPGLAKI FGKDLNLGRP SAAPVIGVPK GDERSWEQRA 300 AGAGVGSSLL PNVQKGLQNF TWSQARGLNS HQQKFGNGIL IVSNEVPHNN GAADSPTTAQ 360 FQLQKATQLQ KLPQLQKQPP QLVKPLQMVN QQQLQAQAPR QIDFSAGTSS KSGVLVARTA 420 VLDGESSEVN GLCKEEGAPP IIEDRRPRKR GRKPANGREE PLNHVEAERQ RREKLNQRFY 480 ALRAVVPNIS KMDKASLLGD AIAYITDLQK KLKDMETERE RFLESGMADP RDRAPRPEVD 540 IQVVRDEVLV RVMSPMENHP VKKVFEAFEE AEVRVGESKV TGNNGTVVHS FIIKCPGSEQ 600 QTREKVIAAM SRAMNMV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 2e-28 | 457 | 519 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 2e-28 | 457 | 519 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 2e-28 | 457 | 519 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 2e-28 | 457 | 519 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 2e-28 | 457 | 519 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 2e-28 | 457 | 519 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 2e-28 | 457 | 519 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 2e-28 | 457 | 519 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 444 | 452 | RRPRKRGRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
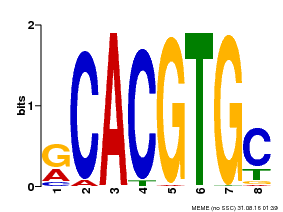
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi2g08080.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK376820 | 0.0 | AK376820.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv3137B23. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003566142.1 | 0.0 | transcription factor bHLH13 | ||||
| TrEMBL | I1HDN7 | 0.0 | I1HDN7_BRADI; Uncharacterized protein | ||||
| STRING | BRADI2G08080.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5174 | 38 | 60 | Representative plant | OGRP2531 | 11 | 23 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 2e-68 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi2g08080.1.p |
| Entrez Gene | 100844558 |



