 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi2g11100.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 482aa MW: 52691 Da PI: 7.8893 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.9 | 5.6e-16 | 351 | 397 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
Bradi2g11100.2.p 351 VHNLSERRRRDRINEKMRALQELIPNC-----NKIDKASMLDEAIEYLKTLQ 397
5*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 1.61E-17 | 344 | 401 | No hit | No description |
| SuperFamily | SSF47459 | 1.44E-20 | 345 | 406 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.981 | 347 | 396 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.9E-13 | 351 | 397 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.6E-20 | 351 | 405 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-17 | 353 | 402 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 482 aa Download sequence Send to blast |
MSDGNEFAEL LWENGQAVVH GRRKHPQPSF QPFGTGSSGV QEKHPGGAGD NMMAFTKAAG 60 VFGGMGIHDL ASGLQHENGD DDAVPWIHYP IMEDDNNNNA PALTTADYSS DFFSELQEAA 120 ANLGSLPPSN HSTNNNRGTP VAGSSRAASK EIQGLSALTT RTAEPQAELA AAKQPRPSDS 180 VMNFSLFSRP AALARATLQS AHRPQGTDKV PNAAANNRME STVIQTTSGP RTGPVFVDQR 240 AAWPQQKEVR FAPAPALTAS VGNLQQEMAR DKLSNNRVVH KDAARKAPDA TVTTSSVCSG 300 NGIVNDEPWH QQKRKIQAEC SASQDEDLDD ESGALLRSTN RSMKRSRTAE VHNLSERRRR 360 DRINEKMRAL QELIPNCNKI DKASMLDEAI EYLKTLQLQV QMMSMGTGLC IPPMLLPPAM 420 QHLQLSQMAH FPHLGMGLGC SRTRSWDPWR KCHANVWNPW ASNPFVRFQY TTIYIFGRSS 480 C* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 355 | 360 | ERRRRD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may act as negative regulator of phyB-dependent light signal transduction. {ECO:0000269|PubMed:17485859}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
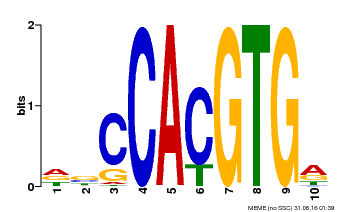
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi2g11100.2.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by light in dark-grown etiolated seedlings. {ECO:0000269|PubMed:17485859}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK102252 | 1e-113 | AK102252.1 Oryza sativa Japonica Group cDNA clone:J033088H11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010233063.2 | 0.0 | transcription factor APG | ||||
| Refseq | XP_024314974.1 | 0.0 | transcription factor APG | ||||
| Swissprot | Q0JNI9 | 1e-178 | PIL15_ORYSJ; Transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15 | ||||
| TrEMBL | A0A2K2D7W8 | 0.0 | A0A2K2D7W8_BRADI; Uncharacterized protein | ||||
| STRING | BRADI2G11100.1 | 0.0 | (Brachypodium distachyon) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 8e-50 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi2g11100.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



