 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi3g10730.1.p | ||||||||
| Common Name | BRADI_3g10730, LOC100841245 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 397aa MW: 43316.3 Da PI: 7.3445 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.8 | 9.2e-32 | 230 | 285 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k+r++W+peLH+rF++a++qLGGs++AtPk+i+elmkv+gLt+++vkSHLQkYRl+
Bradi3g10730.1.p 230 KARRCWAPELHRRFLQALQQLGGSHVATPKQIRELMKVDGLTNDEVKSHLQKYRLH 285
68****************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.868 | 227 | 287 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-27 | 228 | 288 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.87E-16 | 228 | 287 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.8E-27 | 230 | 285 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.7E-6 | 232 | 283 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0055062 | Biological Process | phosphate ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 397 aa Download sequence Send to blast |
MQMAMEEDPA SDTRRRFRDY LLALEEERRK IHVFQRELPL CLDLVTQTIE GMKSQMDAVV 60 GSEETVSDHG PVLEEFIPLK PSLSLCSSEE ESTHAVAPVN SPKKDESERA RPPTPETKKA 120 MPDWLQSVQL WSQEPQQSCP NKELPCKPVA LNATKTRGAF HPFEKEKRAE PELPASSTTA 180 AASSAVVGDS CGDKATSDTE VAHEKIKDTN KDMEKDSKEG QSSQQQHNRK ARRCWAPELH 240 RRFLQALQQL GGSHVATPKQ IRELMKVDGL TNDEVKSHLQ KYRLHTTRRP SSTVAQSNNA 300 AQAAAPQFVV VGSCIWVPPQ EYAAAAAQQQ ANNNSPAVVY APVATMPSSS SALHVPSKKK 360 QTTSRGSKHS EDQRSGDASS SSGSPAVSSS SQTTSA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 2e-13 | 230 | 283 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 2e-13 | 230 | 283 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 2e-13 | 230 | 283 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 2e-13 | 230 | 283 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 2e-13 | 230 | 283 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 2e-13 | 230 | 283 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 2e-13 | 230 | 283 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 2e-13 | 230 | 283 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
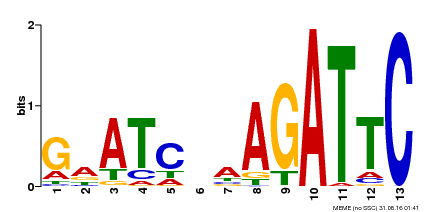
| UniProt | Transcriptional repressor that may play a role in response to nitrogen. May be involved in a time-dependent signaling for transcriptional regulation of nitrate-responsive genes. Binds specifically to the DNA sequence motif 5'-GAATC-3' or 5'-GAATATTC-3'. Represses the activity of its own promoter trough binding to these motifs. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00218 | DAP | Transfer from AT1G68670 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi3g10730.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by nitrate. {ECO:0000269|PubMed:23324170}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003572723.1 | 0.0 | transcription factor NIGT1 | ||||
| Swissprot | Q6Z869 | 1e-123 | NIGT1_ORYSJ; Transcription factor NIGT1 | ||||
| TrEMBL | I1HZP0 | 0.0 | I1HZP0_BRADI; Uncharacterized protein | ||||
| STRING | BRADI3G10730.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6470 | 37 | 54 | Representative plant | OGRP5415 | 12 | 21 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25550.1 | 1e-68 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi3g10730.1.p |
| Entrez Gene | 100841245 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



