 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi3g38200.1.p | ||||||||
| Common Name | BRADI_3g38200, LOC100836269 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 354aa MW: 37489.5 Da PI: 6.821 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 46.1 | 1e-14 | 275 | 327 | 5 | 57 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkeva 57
+r+rr++kNRe+A rsR+RK+a++ eLe +v++L+++N++L+k+ ee+ ++ +
Bradi3g38200.1.p 275 RRQRRMIKNRESAARSRARKQAYTMELEAEVQKLKEQNEELQKKQEEMLEMQK 327
79***************************************999888777655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 2.7E-14 | 270 | 327 | No hit | No description |
| SMART | SM00338 | 5.4E-13 | 271 | 337 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.461 | 273 | 322 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 1.8E-12 | 275 | 328 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 5.88E-11 | 275 | 324 | No hit | No description |
| CDD | cd14707 | 1.09E-27 | 275 | 329 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 278 | 293 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0010255 | Biological Process | glucose mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MDLGDGGESE RKGAAPPMPL ARQGSVYSLT FDEFQSTLGG ASGGGGLGKD FGSMNMDELL 60 RSIWTAEESQ AMASASAAPA GELQRQGSLT LPRTLSIKTV DEVWRDFVRD ASPGAAGGGE 120 PLPKRQPTLG EMTLEDFLVR AGVVRENPAA AAAVDAAVPP PLAARPIQVV NNGSMFFENF 180 GGANGASGAS AMGFAPVGIG DPSHPTMGNG MMPGVAGMGV GAVTVGPLDT SMGQLDSVGK 240 VNGELSSPVE PVPYPFEGVI RGRRSGGHVE KVVERRQRRM IKNRESAARS RARKQAYTME 300 LEAEVQKLKE QNEELQKKQE EMLEMQKNKA LEVINNPYGH KKRCLRRTLT GPW* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that mediates abscisic acid (ABA) signaling. Binds specifically to the ABA-responsive element (ABRE) of the EMP1 and RAB16A gene promoters. {ECO:0000269|PubMed:10611387}. | |||||
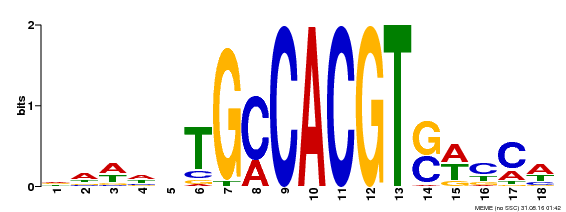
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00186 | DAP | Transfer from AT1G45249 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi3g38200.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By abscisic acid (ABA). {ECO:0000269|PubMed:10611387}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ850323 | 0.0 | KJ850323.1 Brachypodium distachyon ABF2 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001304807.1 | 0.0 | bZIP transcription factor TRAB1 | ||||
| Swissprot | Q6ZDF3 | 1e-150 | TRAB1_ORYSJ; bZIP transcription factor TRAB1 | ||||
| TrEMBL | I1I7U3 | 0.0 | I1I7U3_BRADI; ABF2 | ||||
| STRING | BRADI3G38200.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP706 | 38 | 147 | Representative plant | OGRP540 | 15 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G19290.1 | 2e-27 | ABRE binding factor 4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi3g38200.1.p |
| Entrez Gene | 100836269 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



