 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi3g56290.1.p | ||||||||
| Common Name | BRADI_3g56290, LOC100826037 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 171aa MW: 19593.1 Da PI: 8.4706 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 40.5 | 6.2e-13 | 79 | 137 | 4 | 62 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevaklkse 62
+r rr+ +NRe+ArrsR RK++++ eL v L + N++L ++l++ +++ +
Bradi3g56290.1.p 79 ERRKRRMLSNRESARRSRMRKQKQLSELWAQVVHLRSTNRQLLDQLNHVIRDCDRILHD 137
6899******************************************9998888876655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 2.8E-12 | 76 | 154 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 3.5E-10 | 76 | 133 | No hit | No description |
| PROSITE profile | PS50217 | 10.53 | 78 | 141 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 3.1E-10 | 78 | 136 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 6.88E-12 | 80 | 130 | No hit | No description |
| CDD | cd14702 | 5.65E-16 | 81 | 132 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 83 | 98 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 171 aa Download sequence Send to blast |
MYPAELASIP YLSSASAASF KPHYQVATND ILFQYNSLPV PQAISYQHVE HLVHEASLPV 60 GNKSNSDESD DYQHSLAEER RKRRMLSNRE SARRSRMRKQ KQLSELWAQV VHLRSTNRQL 120 LDQLNHVIRD CDRILHDNSK LRAEQAELKQ QLEKLPVENM ESSVMGPGMT * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 79 | 100 | RRKRRMLSNRESARRSRMRKQK |
| 2 | 92 | 99 | RRSRMRKQ |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00647 | PBM | Transfer from LOC_Os02g49560 | Download |
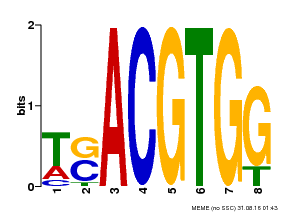
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi3g56290.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003570460.1 | 1e-123 | basic leucine zipper 43 | ||||
| TrEMBL | I1IEA4 | 1e-122 | I1IEA4_BRADI; Uncharacterized protein | ||||
| STRING | BRADI3G56290.1 | 1e-123 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1186 | 35 | 126 | Representative plant | OGRP3104 | 11 | 29 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G30530.1 | 4e-27 | basic leucine-zipper 42 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi3g56290.1.p |
| Entrez Gene | 100826037 |



