 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi4g01730.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 887aa MW: 98252.9 Da PI: 6.0432 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.6 | 2.4e-23 | 122 | 222 | 1 | 98 |
EEEE-..-HHHHTT-EE--HHH.HTT......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr 87
f+k+lt sd++++g +++p++ ae+ +++++++ +l+ +d++ ++W++++i+r++++r++lt+GW+ Fv+a++L +gD+v+F ++
Bradi4g01730.1.p 122 FCKTLTASDTSTHGGFSVPRRSAEKVfppldfSLQPPCQ-ELIARDLHDNEWKFRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVIFI--WN 211
99******************************9999996.9************************************************..67 PP
SEE..EEEEE- CS
B3 88 sefelvvkvfr 98
++ +l+++++
Bradi4g01730.1.p 212 DNNQLLLGIRH 222
67777888875 PP
| |||||||
| 2 | Auxin_resp | 116.5 | 2.3e-38 | 248 | 331 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaa+t+s F+++YnPr+s+seFv++++k++k+++ ++vsvGmRf+m fete+ss rr++Gtv+++sdld+vrWpnS+Wrs+k
Bradi4g01730.1.p 248 AAHAAATNSRFTIFYNPRSSPSEFVIPLAKYVKSVYhTRVSVGMRFRMLFETEESSVRRYMGTVTAISDLDSVRWPNSHWRSVK 331
79*********************************9789******************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 1.09E-47 | 109 | 251 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 1.4E-40 | 113 | 237 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 6.62E-22 | 121 | 214 | No hit | No description |
| Pfam | PF02362 | 2.3E-21 | 122 | 222 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.9E-23 | 122 | 224 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.111 | 122 | 224 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 4.0E-33 | 248 | 331 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 2.7E-8 | 750 | 838 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 1.67E-6 | 752 | 828 | No hit | No description |
| PROSITE profile | PS51745 | 23.91 | 753 | 837 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 887 aa Download sequence Send to blast |
MTQTLPENDG EQRCLNSELW HACAGPLVSL PVVGSRVIYF PQGHSEQVAA STNKEVDGQI 60 PNYPNLPPQL ICQLHNVTMH ADVETDEVYA QMTLQPLSPE EQKEPFLPIE LGAASKQPTN 120 YFCKTLTASD TSTHGGFSVP RRSAEKVFPP LDFSLQPPCQ ELIARDLHDN EWKFRHIFRG 180 QPKRHLLTTG WSVFVSAKRL VAGDSVIFIW NDNNQLLLGI RHANRPQTIM PSSVLSSDSM 240 HIGLLAAAAH AAATNSRFTI FYNPRSSPSE FVIPLAKYVK SVYHTRVSVG MRFRMLFETE 300 ESSVRRYMGT VTAISDLDSV RWPNSHWRSV KVGWDESTAG EKQPRVSLWE IEPLTTFPMY 360 PTAFPLRLKR PWASGLPSMH GMFNGVKNDD FARYSSLMWL GNGDRGTQSS NFQGLGVSPW 420 LQPRIESPLL GLKPDTYQQM AAAALEEIRA GDPLIQSSAL LQFQQTQNLN GGLDSPYANH 480 VLQQMQYQSS LPTVQEGYNQ YSGNSGFLQS HLQQLQLHNP QQLQKQQELP SLQQQRHQIL 540 QQQSHQEMQQ QLSSSCHRVT DVDSSMPGSE SASQSHSPFY QQNLLEGNND PSLHMHNSFR 600 DFSSQEASNL VSLPQSSQLM APEGWPSKRL AVEPLAHVES RSARPKLENG NHQNSISHFA 660 GTLASESARD CSSVQACGSN IDNQLLSSSL HDGMSSVRGG SGNGTVSMAI PLFRYDGEDL 720 PPANSLATSS CLGESATFNS LDNICGVNPS QGGTFVKVYK SGSPGRSLDI TKFSSYYELR 780 SELEHLFGLE GQLEDPVRSG WQLVFVDREN DILLVGDDPW QEFVNSVWCI KILSPQDVHQ 840 MVRNGEGLLS ASGARMMQGN VCDDYSASHD LQNLTGNIAS VPPLDY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 1e-159 | 3 | 352 | 38 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
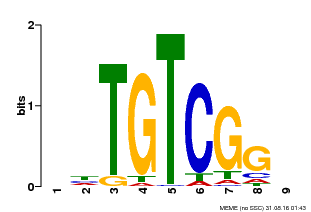
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00033 | PBM | Transfer from AT5G37020 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi4g01730.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK368822 | 0.0 | AK368822.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2080I14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010237021.1 | 0.0 | auxin response factor 25 | ||||
| Swissprot | Q2QM84 | 0.0 | ARFY_ORYSJ; Auxin response factor 25 | ||||
| TrEMBL | A0A0Q3IHY2 | 0.0 | A0A0Q3IHY2_BRADI; Auxin response factor | ||||
| STRING | BRADI4G01730.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP12712 | 29 | 34 | Representative plant | OGRP856 | 14 | 56 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G30330.1 | 0.0 | auxin response factor 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi4g01730.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



