 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi4g26180.1.p | ||||||||
| Common Name | BRADI_4g26180, LOC100830968 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 264aa MW: 28858.5 Da PI: 6.4995 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56 | 9.1e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+ lv +v+++G+g+W+++++ g+ R+ k+c++rw +yl
Bradi4g26180.1.p 14 KGPWTPEEDLVLVSYVQEHGPGNWRSVPASTGLARCSKSCRLRWTNYL 61
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 47.9 | 3.1e-15 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T++Ed ++++ +lG++ W++Ia +++ +Rt++++k++w+++l
Bradi4g26180.1.p 67 RGSFTPQEDAVIAHLQSLLGNR-WAAIATYLP-KRTDNDIKNYWNTHL 112
89********************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-23 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 25.418 | 9 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.09E-30 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.8E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.92E-12 | 16 | 61 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.7E-24 | 65 | 116 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.9 | 66 | 116 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.5E-14 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.0E-14 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.98E-11 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 264 aa Download sequence Send to blast |
MGRPPCCDKV GVKKGPWTPE EDLVLVSYVQ EHGPGNWRSV PASTGLARCS KSCRLRWTNY 60 LRPGIRRGSF TPQEDAVIAH LQSLLGNRWA AIATYLPKRT DNDIKNYWNT HLKKRLQKQQ 120 AVGAIFAPPP TESAACSFSS PSSLSYASSV DNISRLLDGF MKASLPPPAS SAPTTQSSQS 180 VSDAKDKPGH AAVHFIDADE YPMFPAFHDD GCCISGSFAS DQQQQVPLSS IEEWLFDEAV 240 EPKMMDQLAS SSDECYSLVP MLF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 2e-28 | 14 | 116 | 4 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1h88_C | 1e-27 | 14 | 116 | 58 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-27 | 14 | 116 | 58 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1mse_C | 3e-28 | 14 | 116 | 4 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 3e-28 | 14 | 116 | 4 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the regulation of gene (e.g. drought-regulated and flavonoid biosynthetic genes) expression and stomatal movements leading to negative regulation of responses to drought and responses to other physiological stimuli (e.g. light). {ECO:0000269|PubMed:22018045}. | |||||
| UniProt | Transcription factor involved in the regulation of gene (e.g. drought-regulated and flavonoid biosynthetic genes) expression and stomatal movements leading to negative regulation of responses to drought and responses to other physiological stimuli (e.g. light) (PubMed:16005291, PubMed:21637967). Promotes guard cell deflation in response to water deficit. Triggers root growth upon osmotic stress (e.g. mannitol containing medium) (PubMed:21637967). {ECO:0000269|PubMed:16005291, ECO:0000269|PubMed:21637967}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00132 | DAP | Transfer from AT1G08810 | Download |
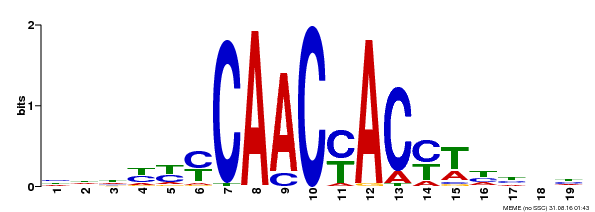
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi4g26180.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by jasmonic acid (JA) and salicylic acid (SA) (PubMed:16463103). Triggered by auxin (IAA) in roots (PubMed:9839469, PubMed:21637967). Stimulated by light, UV-light and cold (PubMed:9839469). According to PubMed:16005291 and PubMed:23828545, rapidly repressed by abscisic acid (ABA) in an ABI1-dependent manner. But in contrast, according to PubMed:21637967, transiently induced by ABA in seedlings (PubMed:16005291, PubMed:21637967, PubMed:23828545). Rapidly repressed by drought. Activated by white light, but repressed by blue light and darkness (PubMed:16005291). Transiently induced by salt (NaCl) in seedlings. Induced by sucrose (PubMed:21637967). Positively regulated by SCAP1 (PubMed:23453954). {ECO:0000269|PubMed:16005291, ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:21637967, ECO:0000269|PubMed:23453954, ECO:0000269|PubMed:23828545, ECO:0000269|PubMed:9839469}. | |||||
| UniProt | INDUCTION: Repressed by abscisic acid (ABA) and osmotic stress (salt stress). {ECO:0000269|PubMed:22018045}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003576343.1 | 0.0 | myb-related protein 306 | ||||
| Swissprot | B3VTV7 | 2e-84 | MYB60_VITVI; Transcription factor MYB60 | ||||
| Swissprot | Q8GYP5 | 7e-85 | MYB60_ARATH; Transcription factor MYB60 | ||||
| TrEMBL | I1INP5 | 0.0 | I1INP5_BRADI; Uncharacterized protein | ||||
| STRING | BRADI4G26180.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP229 | 38 | 296 | Representative plant | OGRP5 | 17 | 1784 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08810.1 | 3e-80 | myb domain protein 60 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi4g26180.1.p |
| Entrez Gene | 100830968 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



