 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi4g33750.1.p | ||||||||
| Common Name | BRADI_4g33750, LOC100824026 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 609aa MW: 67407.6 Da PI: 5.8191 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 424.2 | 1.3e-129 | 35 | 412 | 1 | 349 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLra 93
eel +rmwkd+++l+r+ker+++l ++a+++++k + +++qa rkkm+ra DgiLkYMlk mevcna+GfvYgiip+kgkpv+gasd++ra
Bradi4g33750.1.p 35 EELARRMWKDRVRLRRIKERQQKLA-LQQAELEKSKPKPISDQAMRKKMARAHDGILKYMLKLMEVCNARGFVYGIIPDKGKPVSGASDNIRA 126
8********************9865.66777************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT- CS
EIN3 94 WWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelgl 186
WWkekv+fd+ngpaai+ky+a+nl++++++s + ++ hsl++lqD+tlgSLLs+lmqhc+ppqr++plekg++pPWWP+G+e+ww +lgl
Bradi4g33750.1.p 127 WWKEKVKFDKNGPAAIAKYDAENLVAADAQST---AVKNDHSLMDLQDATLGSLLSSLMQHCSPPQRKYPLEKGTPPPWWPSGDEEWWIALGL 216
*************************9987777...78******************************************************** PP
-TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX......XX..XXXXXX CS
EIN3 187 skdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah......ss..slrkqs 271
+ q pykkphdlkk+wkv+vLt+vikhmsp++++ir+++r+sk+lqdkm+akes ++l vl++ee++++++ + ss + ++
Bradi4g33750.1.p 217 PSGQI-APYKKPHDLKKVWKVGVLTCVIKHMSPNFDKIRNHVRKSKCLQDKMTAKESLIWLGVLQREERLVHSIGNSvlaitySSapE-YRNV 307
**999.8****************************************************************99988865443311433.3366 PP
XXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX..............................XXXXXXXXXXXXXXXXXXXXX......XXXXX CS
EIN3 272 pkvtlsceqkedvegkkeskikhvqavktta..............................gfpvvrkrkkkpsesakvsskevsrtcqssqf 334
+ t s+++++dv+g +e ++++++++++ + + +++++ ++a ss+ + q+
Bradi4g33750.1.p 308 NGNTNSSSNEYDVDGFEEAPLSASSKDDEQDlppaaqsseehvslrgrerantkhpnqavlK---AGAKEQPNIKRARHSSTAIEHGVQRTDD 397
667778888*****999999999999999889999999888888888887777777664331...2233334444444444444444444444 PP
XX.XXXXXXXXXXXX CS
EIN3 335 rgsetelifadknsi 349
+++++ ++d+n++
Bradi4g33750.1.p 398 APENSRNLIPDMNRL 412
445666666666665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.2E-124 | 35 | 283 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 3.0E-68 | 157 | 289 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 9.94E-59 | 164 | 286 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 609 aa Download sequence Send to blast |
MDQLAMLATE FGDSSDFEVD GITENDVSDE EIEPEELARR MWKDRVRLRR IKERQQKLAL 60 QQAELEKSKP KPISDQAMRK KMARAHDGIL KYMLKLMEVC NARGFVYGII PDKGKPVSGA 120 SDNIRAWWKE KVKFDKNGPA AIAKYDAENL VAADAQSTAV KNDHSLMDLQ DATLGSLLSS 180 LMQHCSPPQR KYPLEKGTPP PWWPSGDEEW WIALGLPSGQ IAPYKKPHDL KKVWKVGVLT 240 CVIKHMSPNF DKIRNHVRKS KCLQDKMTAK ESLIWLGVLQ REERLVHSIG NSVLAITYSS 300 APEYRNVNGN TNSSSNEYDV DGFEEAPLSA SSKDDEQDLP PAAQSSEEHV SLRGRERANT 360 KHPNQAVLKA GAKEQPNIKR ARHSSTAIEH GVQRTDDAPE NSRNLIPDMN RLDHVEIPGM 420 SNQIISFTQV GVTSESLQHR GDQGHVYLPG GGANSFDNAE AANATPVSIY MGGQPVPYES 480 SDNARSKSGN PFPLADDSGF NNLPSSYKTP LKQSLPLSRM DHHVVPAGIR APAVNSPYGD 540 HVIDGGNSTS VPGDMQHLID FPFYPEQDKF VGSSFEGLPL DYISISSPIP DIDDFLLHDD 600 DLMEYLGT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 4e-64 | 166 | 292 | 12 | 138 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
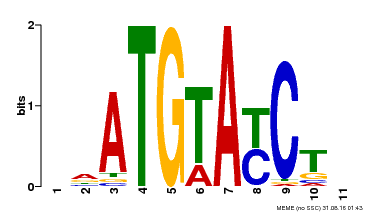
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi4g33750.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024318458.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_024318459.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-141 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | I1IR95 | 0.0 | I1IR95_BRADI; Uncharacterized protein | ||||
| STRING | BRADI4G33750.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3720 | 37 | 79 | Representative plant | OGRP563 | 16 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-138 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi4g33750.1.p |
| Entrez Gene | 100824026 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



