 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bradi5g08980.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 381aa MW: 41297.7 Da PI: 8.8124 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 63.7 | 4.1e-20 | 115 | 204 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkm....rergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
+Wt+ + L++a++e+ e+l+rg+l++ +We+v++++ ++g +s +qCk+k++nl+kryk + +++++ + s+s++p+++q+e
Bradi5g08980.2.p 115 EWTDGAISSLLDAYTERFEQLNRGNLRGRDWEDVAAAVtdgqGKGGAGKSVEQCKNKIDNLKKRYKVECTRNGGAGAASVSHWPWYRQME 204
5*************************************9999999****************************9999999*********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF13837 | 3.7E-21 | 113 | 206 | No hit | No description |
| Gene3D | G3DSA:3.40.1160.10 | 2.1E-7 | 258 | 303 | IPR001048 | Aspartate/glutamate/uridylate kinase |
| SuperFamily | SSF53633 | 9.16E-5 | 260 | 303 | IPR001048 | Aspartate/glutamate/uridylate kinase |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006221 | Biological Process | pyrimidine nucleotide biosynthetic process | ||||
| GO:0046939 | Biological Process | nucleotide phosphorylation | ||||
| GO:0005737 | Cellular Component | cytoplasm | ||||
| GO:0033862 | Molecular Function | UMP kinase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 381 aa Download sequence Send to blast |
MASCDDDFGL LGDDAHPAPQ PSAQPPPPQQ AQTFCFVDAP ATGSGSGPFV PAQEEGNHSS 60 ERGKASHHSK RRRERAEEFS DGGEYCSYIS SSAGGRKGRG GGGGGSSDYR KDREEWTDGA 120 ISSLLDAYTE RFEQLNRGNL RGRDWEDVAA AVTDGQGKGG AGKSVEQCKN KIDNLKKRYK 180 VECTRNGGAG AASVSHWPWY RQMEQIIGNS SSPGTSKPLA PTNDEKPRQL LQNANKRYPS 240 SGTGPPTMVP CSRLTPLSNP KWKRVLLKIG GTALAGEAPH NVDPKVIMLI AREVQVACRH 300 GVENDGSSDE CSIASSITRK NRCGDTSPDC TDDARGSRTI HKKASHAPSS KRKGSYLWWN 360 WCLHRKSTFH NRYRCCTESF * |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 93 | 101 | GGRKGRGGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00340 | DAP | Transfer from AT3G10030 | Download |
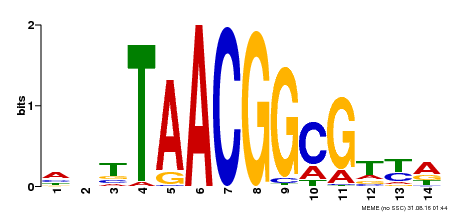
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Bradi5g08980.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK369935 | 0.0 | AK369935.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2101G10. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003581179.1 | 0.0 | uncharacterized protein LOC100840998 isoform X1 | ||||
| TrEMBL | A0A2K2CG48 | 0.0 | A0A2K2CG48_BRADI; Uncharacterized protein | ||||
| STRING | BRADI5G08980.1 | 0.0 | (Brachypodium distachyon) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G10030.2 | 1e-63 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Bradi5g08980.2.p |



