 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Brast01G176300.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 374aa MW: 39886 Da PI: 9.9557 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 37.8 | 4.9e-12 | 63 | 111 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ r +r++lg+f + +Aa+a++aa+++++g
Brast01G176300.1.p 63 SRYKGVVPQP-NGRWGAQIYE------RhQRVWLGTFAAESDAARAYDAAAQRFRG 111
78****9888.8*********......44**********88*************98 PP
| |||||||
| 2 | B3 | 97.4 | 9.1e-31 | 195 | 288 | 1 | 84 |
EEEE-..-HHHHTT-EE--HHH.HTT..........---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh..........ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvv 81
f+k+ tpsdv+k++rlv+pk++ae+h gg++ ++ l +ed g++W+++++y+++s++yvltkGW++Fvk++gL++gD+v
Brast01G176300.1.p 195 FDKTVTPSDVGKLNRLVIPKQHAEKHfplqllpaagGGESGKGLLLNFEDAGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKGLRAGDVVG 285
89*********************************98889999************************************************ PP
EEE CS
B3 82 Fkl 84
F++
Brast01G176300.1.p 286 FYR 288
*93 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.22E-12 | 63 | 119 | No hit | No description |
| Pfam | PF00847 | 2.1E-7 | 63 | 111 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 4.84E-16 | 63 | 119 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 9.9E-27 | 64 | 125 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.796 | 64 | 119 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 9.4E-20 | 64 | 119 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 1.4E-39 | 187 | 318 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.11E-28 | 193 | 288 | No hit | No description |
| SuperFamily | SSF101936 | 2.35E-29 | 193 | 288 | IPR015300 | DNA-binding pseudobarrel domain |
| PROSITE profile | PS50863 | 13.465 | 195 | 307 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.6E-21 | 195 | 306 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 4.6E-27 | 195 | 291 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048527 | Biological Process | lateral root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 374 aa Download sequence Send to blast |
MDSSSCLADE TTSGGASTDK LKALATAATA PLERMGSGAS AVLDAAEPGA EADSGRAGKL 60 PSSRYKGVVP QPNGRWGAQI YERHQRVWLG TFAAESDAAR AYDAAAQRFR GRDAVTNFRP 120 LSEDAAELRF LASRSKAEVV DMLRKHTYFD ELAQSSRAFA AAASVPNIAS RHDGHSSSLS 180 PSPAAAAAAA REHLFDKTVT PSDVGKLNRL VIPKQHAEKH FPLQLLPAAG GGESGKGLLL 240 NFEDAGGKVW RFRYSYWNSS QSYVLTKGWS RFVKEKGLRA GDVVGFYRAV AVAGEDGKLF 300 IDCKLRPDTN TAAAVDQSTP VAKPVRLFGV DLTVRPAPEQ GTAGCKRTTP RDFVEPPGPR 360 VPFKKQCVKL ALV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5os9_A | 7e-51 | 189 | 307 | 2 | 114 | B3 domain-containing transcription factor NGA1 |
| 5os9_B | 7e-51 | 189 | 307 | 2 | 114 | B3 domain-containing transcription factor NGA1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00139 | DAP | Transfer from AT1G13260 | Download |
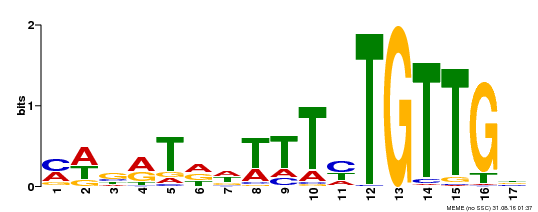
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Brast01G176300.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003569603.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os01g0693400 | ||||
| Swissprot | Q8RZX9 | 1e-176 | Y1934_ORYSJ; AP2/ERF and B3 domain-containing protein Os01g0693400 | ||||
| TrEMBL | I1HQL3 | 0.0 | I1HQL3_BRADI; Uncharacterized protein | ||||
| STRING | BRADI2G47220.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 1e-102 | related to ABI3/VP1 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Brast01G176300.1.p |



