 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Brast01G210100.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 339aa MW: 36928.1 Da PI: 8.3288 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 102.3 | 2.8e-32 | 193 | 250 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s++prsYYrCt+++C vkk+vers +dp++v++tYeg+H+h+
Brast01G210100.1.p 193 EDGYRWRKYGQKAVKNSPYPRSYYRCTTPKCGVKKRVERSYQDPSTVITTYEGQHTHH 250
8********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.5E-33 | 179 | 252 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 6.67E-29 | 185 | 252 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 31.066 | 187 | 252 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.6E-37 | 192 | 251 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 1.9E-25 | 193 | 250 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 339 aa Download sequence Send to blast |
MSSGGGGGGG DRHGHGLYHL GSDYELSSND LESFFFSQPA AGDIGGGVGA EEIAPYASIT 60 DYLQGILDPS TGIARHLDVP CSSPVKHELS VEARSHDLDS QGAGSAAGAL LTPNSSLSFS 120 SSGGDDGEGK SRRCKKQQGR AKKAEEDQGG KDLQDDGENS KKGNSKAKKQ KAEKRQRLPR 180 VSFLTKSEVD HLEDGYRWRK YGQKAVKNSP YPRSYYRCTT PKCGVKKRVE RSYQDPSTVI 240 TTYEGQHTHH SPASLRGSAA HLFMPPPQHL GLMAPPLFRP DLMSMMQHMQ YSNPNNMYMP 300 TSIPPHHHPM ATPPLQQHHF TDYALLQDLF PSTMPNNP* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 2e-29 | 185 | 253 | 10 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 2e-29 | 185 | 253 | 10 | 78 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
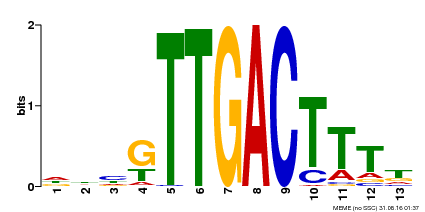
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00440 | DAP | Transfer from AT4G18170 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Brast01G210100.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU665451 | 1e-130 | EU665451.1 Triticum aestivum WRKY36 transcription factor mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003566905.1 | 0.0 | probable WRKY transcription factor 71 | ||||
| Swissprot | Q8VWJ2 | 8e-49 | WRK28_ARATH; WRKY transcription factor 28 | ||||
| TrEMBL | I1HPL1 | 0.0 | I1HPL1_BRADI; Uncharacterized protein | ||||
| STRING | BRADI2G44090.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP623 | 37 | 173 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G29860.1 | 4e-48 | WRKY DNA-binding protein 71 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Brast01G210100.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



