 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Brast01G348100.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 268aa MW: 30172.9 Da PI: 9.5508 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 98.5 | 2.8e-31 | 43 | 93 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
krien++nrqvtf+kRrng+lKKA+ELSvLCdaeva+++fss+g+lyeys+
Brast01G348100.1.p 43 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVVFSSRGRLYEYSN 93
79***********************************************95 PP
| |||||||
| 2 | K-box | 92.3 | 8.2e-31 | 111 | 208 | 4 | 100 |
K-box 4 ssgks.leeakaeslqqelakLkkeienLq.reqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenk 92
s++ ++e +a+++qqe++kL+++i++Lq ++ R l+ +++++++l++L+qLe++Lek++ kiR++Knell++++e++qk+e el+++n+
Brast01G348100.1.p 111 SNS-GtVAEVNAQHYQQESSKLRQQISSLQnSNSRSLVKDSVSTMTLRDLKQLESRLEKGIAKIRARKNELLYAEVEYMQKREMELHNDNM 200
233.25999*********************9899********************************************************* PP
K-box 93 aLrkklee 100
+Lr+k++e
Brast01G348100.1.p 201 YLRTKVAE 208
*****986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 33.711 | 35 | 95 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 3.6E-41 | 35 | 94 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 2.09E-32 | 36 | 107 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 8.25E-45 | 36 | 109 | No hit | No description |
| PRINTS | PR00404 | 2.1E-32 | 37 | 57 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 37 | 91 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.2E-26 | 44 | 91 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.1E-32 | 57 | 72 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.1E-32 | 72 | 93 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 8.5E-22 | 120 | 206 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 13.78 | 121 | 212 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 268 aa Download sequence Send to blast |
MMNMMGDLSC GGASSGKDQP GGTASAGEAA GSEKMGRGRI EIKRIENTTN RQVTFCKRRN 60 GLLKKAYELS VLCDAEVALV VFSSRGRLYE YSNNSVKATI ERYKKANSDT SNSGTVAEVN 120 AQHYQQESSK LRQQISSLQN SNSRSLVKDS VSTMTLRDLK QLESRLEKGI AKIRARKNEL 180 LYAEVEYMQK REMELHNDNM YLRTKVAENE RGQQPMNMMA AASTSSEYDH MVQYDSRNFL 240 QVNLMQQQQY SHQLQPTALQ LGQQHFN* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 1e-20 | 35 | 122 | 1 | 91 | MEF2C |
| 5f28_B | 1e-20 | 35 | 122 | 1 | 91 | MEF2C |
| 5f28_C | 1e-20 | 35 | 122 | 1 | 91 | MEF2C |
| 5f28_D | 1e-20 | 35 | 122 | 1 | 91 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in the development of floral organs. Acts as C-class protein in association with MADS58. Involved in the control of lodicule number (whorl 2), stamen specification (whorl 3) and floral meristem determinacy (whorl 4), but not in the regulation of carpel morphogenesis. Plays a more predominant role in controlling lodicule development and in specifying stamen identity than MADS58. {ECO:0000269|PubMed:11828031, ECO:0000269|PubMed:16326928, ECO:0000269|PubMed:9869408}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00609 | ChIP-seq | Transfer from AT4G18960 | Download |
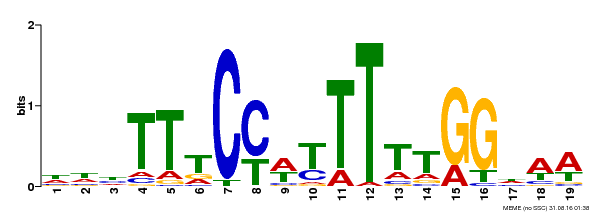
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Brast01G348100.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KF469309 | 0.0 | KF469309.1 Brachypodium distachyon MADS-box transcription factor 14 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001289811.1 | 1e-172 | MADS-box transcription factor 3 | ||||
| Refseq | XP_010230591.1 | 1e-172 | MADS-box transcription factor 3 isoform X1 | ||||
| Refseq | XP_010230592.1 | 1e-172 | MADS-box transcription factor 3 isoform X1 | ||||
| Swissprot | Q40704 | 1e-149 | MADS3_ORYSJ; MADS-box transcription factor 3 | ||||
| TrEMBL | I1HD30 | 1e-170 | I1HD30_BRADI; MADS-box transcription factor 14 | ||||
| STRING | BRADI2G06330.1 | 1e-171 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP649 | 37 | 144 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18960.1 | 1e-101 | MIKC_MADS family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Brast01G348100.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



