 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Brast02G057700.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 332aa MW: 36510.8 Da PI: 6.6184 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56.1 | 6.2e-18 | 113 | 166 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rqV +WFqNrRa++k
Brast02G057700.1.p 113 KKRRLNVEQVRTLEKNFELANKLEPERKIQLARALGLQPRQVAIWFQNRRARWK 166
4568999**********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 129.8 | 1.1e-41 | 112 | 202 | 1 | 91 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreel 91
ekkrrl+ eqv++LE++Fe +kLeperK +lar+Lglqprqva+WFqnrRAR+ktkqlEkdy++Lkr++da+k+en++L +++++L++e+
Brast02G057700.1.p 112 EKKRRLNVEQVRTLEKNFELANKLEPERKIQLARALGLQPRQVAIWFQNRRARWKTKQLEKDYDVLKRQFDAVKAENDALISHNKKLQSEI 202
69**************************************************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-19 | 93 | 169 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.69E-19 | 103 | 170 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.086 | 108 | 168 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 6.2E-17 | 111 | 172 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 6.56E-17 | 113 | 169 | No hit | No description |
| Pfam | PF00046 | 2.9E-15 | 113 | 166 | IPR001356 | Homeobox domain |
| PRINTS | PR00031 | 1.7E-5 | 139 | 148 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 143 | 166 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.7E-5 | 148 | 164 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.6E-15 | 168 | 207 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 332 aa Download sequence Send to blast |
MASNGMASSP SAFFPPNFLL HMQQTPPVVH HDPEEHHHHH HEHHLLPPPP HHGGHPHNPF 60 LPPSSLQDFR GMAPMLGKRP AMYGGEGGGE DVNGGGANEE EMSDDGSQAG GEKKRRLNVE 120 QVRTLEKNFE LANKLEPERK IQLARALGLQ PRQVAIWFQN RRARWKTKQL EKDYDVLKRQ 180 FDAVKAENDA LISHNKKLQS EILGLKGCSR EAASELINLN KETEASCSNR SENSSEINLD 240 ISRTPPPSEG GPMDAPPHQN GGGMIPFYPS AVVARPGAGV DIDQLLQASS VPKMEQHHHG 300 GAADTPSFGN LLCGVDEPPP FWPWADHQHF H* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 160 | 168 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
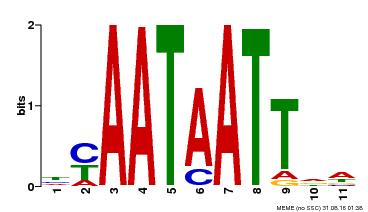
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Brast02G057700.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK359041 | 0.0 | AK359041.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1087P18. | |||
| GenBank | AK359747 | 0.0 | AK359747.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv1102E15. | |||
| GenBank | AK366245 | 0.0 | AK366245.1 Hordeum vulgare subsp. vulgare mRNA for predicted protein, complete cds, clone: NIASHv2041G22. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003558711.1 | 0.0 | homeobox-leucine zipper protein HOX21 | ||||
| Swissprot | Q8S7W9 | 1e-137 | HOX21_ORYSJ; Homeobox-leucine zipper protein HOX21 | ||||
| TrEMBL | I1H976 | 0.0 | I1H976_BRADI; Uncharacterized protein | ||||
| STRING | BRADI1G73190.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2289 | 38 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 1e-80 | HD-ZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Brast02G057700.1.p |



