 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Brast04G314200.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 711aa MW: 79499.8 Da PI: 7.3733 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 154.8 | 6.5e-48 | 101 | 244 | 4 | 144 |
DUF822 4 grkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl..eeaeaagssasaspes 92
g++++++E+E++k+RER+RR i+++++aGLR++Gn++lp+raD+n+Vl+AL+r+AGw+v++DGtt+r + +pl a+ g+ + +s e+
Brast04G314200.1.p 101 GEREREREKERTKLRERHRRSITSRMLAGLRQHGNFPLPARADMNDVLAALARAAGWTVQPDGTTFRSSPQPLlpPPAQFQGALQVTSMET 191
67899*******************************************************************9988899999999999999 PP
DUF822 93 slq.sslkssalaspvesysaspksssfpspssldsislasaasllpvlsvls 144
+ ++l+s+a+ +p++s+ + +++++++spssl+s+ +a+++ + +
Brast04G314200.1.p 192 PALaNALNSYAIGTPLDSQTSALQTDDSLSPSSLNSVVVAEQSIKNENYGNSC 244
998899*********************************99844433333333 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 9.7E-48 | 101 | 251 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 3.93E-163 | 270 | 703 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 6.9E-170 | 270 | 705 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 5.5E-82 | 293 | 665 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.1E-51 | 308 | 322 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.1E-51 | 329 | 347 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.1E-51 | 351 | 372 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.1E-51 | 444 | 466 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.1E-51 | 517 | 536 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.1E-51 | 551 | 567 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.1E-51 | 568 | 579 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.1E-51 | 586 | 609 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 4.1E-51 | 624 | 646 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 711 aa Download sequence Send to blast |
MFGLGQLCYI SIPASGLHSS LRFLAGSVGG RPSSPAALAS ADRRRRLRRA AMSTKHPHAH 60 PRDADPAASP PPAPPSRRAR GGFASSTPAA PAPPATTRRR GEREREREKE RTKLRERHRR 120 SITSRMLAGL RQHGNFPLPA RADMNDVLAA LARAAGWTVQ PDGTTFRSSP QPLLPPPAQF 180 QGALQVTSME TPALANALNS YAIGTPLDSQ TSALQTDDSL SPSSLNSVVV AEQSIKNENY 240 GNSCSANSLN CIGSDQLMRA SAVSAGDYTR TPYIPVYASL PMGIINCYCQ LVDPEALRAE 300 LRHLKSLNVD GVIVDCWWGV VEAWTPQKYE WSGYRDLFGI IKEFRLKVQV VLSFHGSGEC 360 ESGDLLISLP RWVMEIAQEN QDIFFTDREG RRNTECLSWG IDKERVLRGR TGIEVYFDFM 420 RSFHMEFRSL SEEGLVSAIE IGLGASGELR YPACTKKMGW RYPGIGEFQC YDRYMQKNLR 480 QSALKRGHLF WARGPDNAGY YNSRSHETGF FCDGGDYDSY YGRFFLNWYS GILVDHVDQV 540 LSLATLAFDG AEIVVKIPSI YWWYRTASHA AELTAGFYNP TNRDGYSPVF RMLKKHSVIL 600 KLVCYGPEFT VQENDEAFAD PEGLTWQVMN AAWDHGLSLS VESALPCLDG EMYPQILDIA 660 KPRNDPDRHH VSFFAYRQQP PFLLQRYVCF SELETFVKCM HGEDTQNFID * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-119 | 273 | 703 | 11 | 444 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
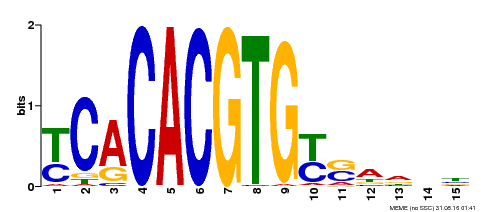
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Brast04G314200.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_014755446.1 | 0.0 | beta-amylase 8 isoform X1 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | I1HWU7 | 0.0 | I1HWU7_BRADI; Beta-amylase | ||||
| STRING | BRADI3G02800.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2141 | 36 | 63 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Brast04G314200.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



