 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Brast05G165200.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 609aa MW: 67376.4 Da PI: 5.6247 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 428.5 | 6.4e-131 | 35 | 413 | 1 | 350 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsL 91
eel +rmwkd+++l+r+ker+++l ++a+++++k + +++qa rkkmsra DgiLkYMlk mevcna+GfvYgiip+kgkpv+gasd++
Brast05G165200.1.p 35 EELARRMWKDRVRLRRIKERQQKLA-LQQAELEKSKPKPISDQAMRKKMSRAHDGILKYMLKLMEVCNARGFVYGIIPDKGKPVSGASDNI 124
8********************9865.66777************************************************************ PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHH CS
EIN3 92 raWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwg 182
raWWkekv+fd+ngpaai+ky+a+nl+++++ s+ + ++ hsl++lqD+tlgSLLs+lmqhc+ppqr++plekg++pPWWP+G+e+ww
Brast05G165200.1.p 125 RAWWKEKVKFDKNGPAAIAKYDAENLVAADAPSS---AVKNEHSLMDLQDATLGSLLSSLMQHCNPPQRKYPLEKGTPPPWWPSGNEDWWI 212
*****************************98888...789*************************************************** PP
HHT--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX......XXXX CS
EIN3 183 elglskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah......sssl 267
+lgl+ q pykkphdlkk+wkv+vLt+vikhmsp++++ir+++r+sk+lqdkm+akes ++l vl++ee++++++ + s +
Brast05G165200.1.p 213 ALGLPSGQI-APYKKPHDLKKVWKVGVLTCVIKHMSPNFDKIRNHVRKSKCLQDKMTAKESLIWLGVLQREERLVHSIGNSvlaityS--S 300
******999.8****************************************************************9999886543321..2 PP
XX...XXXXXXXXXXXXXXXXXXXXXX.XXXXXXXXXX................................XXXXXXXXXXXXXXXXXXXXX CS
EIN3 268 rk...qspkvtlsceqkedvegkkeskikhvqavktta................................gfpvvrkrkkkpsesakvssk 323
+ + t s+++++dv+g +e ++++++++++ g++ krk+ ++s++++++
Brast05G165200.1.p 301 APeygNVNGNTNSSSNEYDVDGFEEAPLSASSKDDEQDlsppaqsseehvsargrerantkhpnqdvlkaGAKERPKRKRACRSSTAIEDE 391
2224466677888999*****999999999999999999*******999****99999999999977777777777777666666666555 PP
......XXXXXXX.XXXXXXXXXXXXX CS
EIN3 324 evsrtcqssqfrgsetelifadknsis 350
q+ +++++ ++d+n+++
Brast05G165200.1.p 392 -----VQRTDDAPENSRNLIPDMNRLD 413
.....4444444456778888888766 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.1E-124 | 35 | 283 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.2E-68 | 157 | 289 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 2.88E-59 | 163 | 286 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 609 aa Download sequence Send to blast |
MDQLALLATE FGDSSDFEVD GITENDVSDE EIEPEELARR MWKDRVRLRR IKERQQKLAL 60 QQAELEKSKP KPISDQAMRK KMSRAHDGIL KYMLKLMEVC NARGFVYGII PDKGKPVSGA 120 SDNIRAWWKE KVKFDKNGPA AIAKYDAENL VAADAPSSAV KNEHSLMDLQ DATLGSLLSS 180 LMQHCNPPQR KYPLEKGTPP PWWPSGNEDW WIALGLPSGQ IAPYKKPHDL KKVWKVGVLT 240 CVIKHMSPNF DKIRNHVRKS KCLQDKMTAK ESLIWLGVLQ REERLVHSIG NSVLAITYSS 300 APEYGNVNGN TNSSSNEYDV DGFEEAPLSA SSKDDEQDLS PPAQSSEEHV SARGRERANT 360 KHPNQDVLKA GAKERPKRKR ACRSSTAIED EVQRTDDAPE NSRNLIPDMN RLDHVEIPGM 420 SNQIISFNQV GVTSGSLQHR GDQGHVYLPG GGANSFDNAQ AANATPVSIY MGGQPVPYES 480 SDNARSKSGN PFPLDDDSGF NNLPGSYQTP LKQSLPLSRM DHHVVPVGIR APAVNSPYGD 540 HVTDGGNSTS VPGDMQHLID FPFYPEQDKF VGSSFEGLPL DYISISSPIP DIDDFLLHDD 600 DLMEYLGT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 1e-64 | 166 | 292 | 12 | 138 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
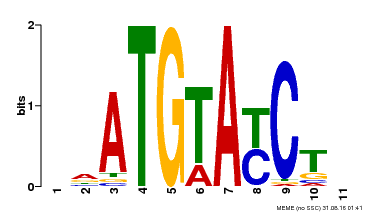
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Brast05G165200.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024318458.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_024318459.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-141 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | I1IR95 | 0.0 | I1IR95_BRADI; Uncharacterized protein | ||||
| STRING | BRADI4G33750.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3720 | 37 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-136 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Brast05G165200.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



