 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Brast08G119600.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 848aa MW: 92968.5 Da PI: 6.2451 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.5 | 5.1e-35 | 164 | 238 | 2 | 76 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
Cqv+gCead+ e+k yh+rh+vC +++a++v+++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
Brast08G119600.1.p 164 CQVPGCEADIRELKGYHKRHRVCLRCAHATAVMLDGVQKRYCQQCGKFHILLDFDEDKRSCRRKLERHNKRRRRK 238
************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 8.2E-27 | 156 | 225 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.359 | 161 | 238 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.19E-33 | 162 | 241 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 8.8E-27 | 164 | 238 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 848 aa Download sequence Send to blast |
MDASDSGGGS AAADAGEPDW DWNNILEFAI RGDDSLTLPW DDTLGTTEAD PAEEAFLPAP 60 SPVLPVEAEP VAPPPPVEAG GSRRGVRKRD PRLVCPNYLA GIVPCACPEL DEMAAAAEVE 120 EVASELLAGP RKKSRPASRG NGVAAGGGGG TGAAGRGGAA EMKCQVPGCE ADIRELKGYH 180 KRHRVCLRCA HATAVMLDGV QKRYCQQCGK FHILLDFDED KRSCRRKLER HNKRRRRKPD 240 SKGTVEKEAD EQLDLSADGS GGCELREENT DGTTCEMVET VRSNKVLDRE TPVGSEDVLS 300 SPTCTHRSLQ NEQSKSIVTF AASVEGCLGT EQENAKLTNS PTHDTKSVYS SSCPTGRISF 360 KLYDWNPAEF PRRLRNQIFE WLSSMPVELE GYIRPGCTIL TVFIAMPQHM WDQLSEDAAN 420 LVRDLVNAPS SLLLGKGAFF VHVNNMIFQV LKDGATLMST RLEVQAPRIH YVHPTWFEAG 480 KPVELLLCGS SLDQPKFRSL LSFDGEYLKH DCCRLMSHET IACVKNAAAL DSQHEIFRIN 540 ITQTKADTHG PGFVEVENII GLSNFVPILF GSKQLCSELE RIQDALCGSD EKYKSVFGEV 600 PGATSDPCGH RELKQTAMSG FLIEIGWLIR KSSPDELKNL LSSTNIKRWT CVLKFLIQND 660 FINVLEIIVK SSDNIIGSEI LSNLERGRLE HHVTTFLGYV RHARNIVGDR AKYEKQTQLE 720 TRWCGDSASN QPNLGISVPF AKENTGAGGE YDLRPTNVEC KEEERMLLAT KAVSHRQCCR 780 PEMNARWLNP TLGAPFPGGA MRTRLVTTVV VAAVLCFAAC VVVFHPDRVG VLAAPVKRYL 840 FSDSPSS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 5e-38 | 163 | 245 | 5 | 87 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 220 | 238 | KRSCRRKLERHNKRRRRKP |
| 2 | 224 | 236 | RRKLERHNKRRRR |
| 3 | 231 | 238 | NKRRRRKP |
| 4 | 232 | 237 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
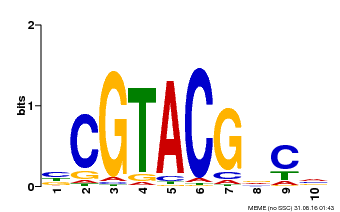
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Brast08G119600.1.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003566250.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | I1HJH8 | 0.0 | I1HJH8_BRADI; Uncharacterized protein | ||||
| STRING | BRADI2G25580.1 | 0.0 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-139 | squamosa promoter binding protein-like 7 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Brast08G119600.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



