 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Brast09G143700.1.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 266aa MW: 29070.3 Da PI: 5.4472 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.9 | 8.3e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT+eEd+ lv +k +G g+W++ ++ g+ R++k+c++rw +yl
Brast09G143700.1.p 14 RGPWTPEEDKTLVAHIKSFGHGNWRALPKQAGLLRCGKSCRLRWINYL 61
89******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 53.6 | 5e-17 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++++eE++ +++++++lG++ W++Ia++++ gRt++++k+ w+++l
Brast09G143700.1.p 67 RGNFSEEEEQTIIQLHELLGNR-WSAIAARLP-GRTDNEIKNVWHTHL 112
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-25 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.952 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.03E-30 | 10 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.1E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.0E-15 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.61E-11 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 26.132 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.4E-27 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.1E-15 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.2E-16 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.63E-12 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 266 aa Download sequence Send to blast |
MGRAPCCEKM GLKRGPWTPE EDKTLVAHIK SFGHGNWRAL PKQAGLLRCG KSCRLRWINY 60 LRPDIKRGNF SEEEEQTIIQ LHELLGNRWS AIAARLPGRT DNEIKNVWHT HLKKRLDPAS 120 ARQEHDEEDE GQASKKRKPA AASRKRKPAT APVSSPERSV STSTLTESSM AAAEHGNSGS 180 SAVSVKEESF TSASEESDEF QIDESFWSET LSMPLLDGGV SMEPDEAFGS GGSASGAVDG 240 DMDYWLRVFM EGGGGDDVAL DLPQI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-28 | 12 | 117 | 5 | 109 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator involved in cold stress response (PubMed:14675437, PubMed:20807373, PubMed:22246661). Regulates positively the expression of genes involved in reactive oxygen species (ROS) scavenging such as peroxidase and superoxide dismutase during cold stress. Transactivates a complex gene network that have major effects on stress tolerance and panicle development (PubMed:20807373). {ECO:0000269|PubMed:14675437, ECO:0000269|PubMed:20807373, ECO:0000269|PubMed:22246661}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00375 | DAP | Transfer from AT3G23250 | Download |
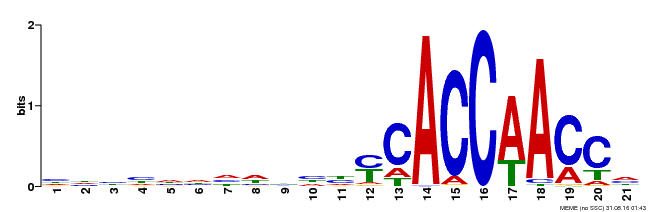
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Brast09G143700.1.p |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold stress. {ECO:0000269|PubMed:14675437, ECO:0000269|PubMed:20807373, ECO:0000269|Ref.2}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY615199 | 1e-145 | AY615199.1 Triticum aestivum transcription factor Myb2 (Myb2) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_003580139.1 | 1e-172 | transcription factor MYB4 | ||||
| Swissprot | Q7XBH4 | 1e-108 | MYB4_ORYSJ; Transcription factor MYB4 | ||||
| TrEMBL | I1IZP3 | 1e-171 | I1IZP3_BRADI; Uncharacterized protein | ||||
| STRING | BRADI5G15760.1 | 1e-172 | (Brachypodium distachyon) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP79 | 38 | 563 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G23250.1 | 9e-81 | myb domain protein 15 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Brast09G143700.1.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



