 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Brast09G253300.2.p | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Pooideae; Brachypodieae; Brachypodium
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 822aa MW: 91557.5 Da PI: 6.2867 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 72.9 | 3.9e-23 | 136 | 237 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkl 84
f+k+lt sd++++g +++p++ ae+ ++++++ +l+ +d + +W++++i+r++++r++lt+GW+ Fv+a++L +gD+v+F
Brast09G253300.2.p 136 FCKTLTASDTSTHGGFSVPRRAAERVfppldftQQPPAQ--ELIARDIHDVEWKFRHIFRGQPKRHLLTTGWSVFVSAKRLVAGDSVLFI- 223
99***********************9*****86555544..8************************************************. PP
-SSSEE..EEEEE-S CS
B3 85 dgrsefelvvkvfrk 99
++++ +l+++++r+
Brast09G253300.2.p 224 -WNEKNQLWLGIRRA 237
.8999999*****97 PP
| |||||||
| 2 | Auxin_resp | 110.4 | 1.8e-36 | 262 | 345 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalk.vkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
aahaast+s F+++YnPra++seFv++++k+ ka+ +++svGmRf+m fete+ss rr++Gt++ vsd+dpvrW S Wrs+k
Brast09G253300.2.p 262 AAHAASTNSRFTIFYNPRACPSEFVIPLSKYIKAVFhTRISVGMRFRMLFETEESSVRRYMGTITEVSDADPVRWASSYWRSVK 345
79*******************************986599******************************************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 7.72E-48 | 121 | 265 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 8.4E-41 | 128 | 251 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.84E-21 | 135 | 236 | No hit | No description |
| Pfam | PF02362 | 1.7E-21 | 136 | 237 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 7.6E-24 | 136 | 238 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 12.083 | 136 | 238 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 1.2E-31 | 262 | 345 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 4.2E-5 | 613 | 809 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 2.03E-7 | 717 | 797 | No hit | No description |
| PROSITE profile | PS51745 | 24.037 | 722 | 806 | IPR000270 | PB1 domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009908 | Biological Process | flower development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 822 aa Download sequence Send to blast |
MSSSSGASVG QQQQPPPPAA PPEEEKKNLN SELWHACAGP LVCLPTVGTR VVYFPQGHSE 60 QVAASTNKEV EGHIPNYPNL PPQLICQLHD VTMHADVETD EVYAQMTLQP LNPQEQNDAY 120 LPAEMGIMSK QPTNYFCKTL TASDTSTHGG FSVPRRAAER VFPPLDFTQQ PPAQELIARD 180 IHDVEWKFRH IFRGQPKRHL LTTGWSVFVS AKRLVAGDSV LFIWNEKNQL WLGIRRASRT 240 QTVMPSSVLS SDSMHIGLLA AAAHAASTNS RFTIFYNPRA CPSEFVIPLS KYIKAVFHTR 300 ISVGMRFRML FETEESSVRR YMGTITEVSD ADPVRWASSY WRSVKVGWDE STAGERPPRV 360 SLWEIEPLTT FPMYPSLFPL RVKHPWYSGV AGLHDDSNAL MWLRGVAGDG GYQSLNFQSP 420 GIGPWGQQRL HPSLMSTDHD QYQAVVAAAA ASQSGGYMKQ QFLNLQQPMQ SPQEHCNLNP 480 LLQQQILQQA SQQQTVSAEN QNIQTMLNSS AIQHQLQQIQ QMQQAHIDQK QKIQPDQTYQ 540 VPTSSALPSP TSLPSHLREK FGFSDPHVNP PTFTTSSSSD NMLESSFLQG SSKAVDLSRF 600 NQPALNEQQQ QQQQQQAWKQ KFMGSPSLSF GGSVLLNSPT SKDGSLENKI GPDGQNQSLF 660 SPQVDSSSLL YNMVPNMTSN VADNNMSTIP SGSTYLQNPM YGCLDDSSGI FQNTGENDPT 720 SRTFVKVYKS GSVGRSLDIT RFSNYAELRE ELGQMFGIRG QLDDPDRSGW QLVFVDREND 780 VLLLGDDPWE SFVNSVWYIK ILSPEDVHKL GKQGNDPRYL S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldu_A | 1e-160 | 25 | 366 | 47 | 388 | Auxin response factor 5 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
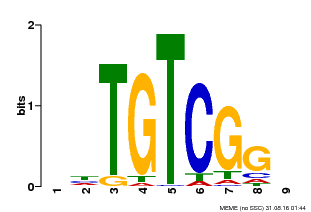
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00033 | PBM | Transfer from AT5G37020 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Brast09G253300.2.p |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK071455 | 0.0 | AK071455.1 Oryza sativa Japonica Group cDNA clone:J023093B04, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_014751484.1 | 0.0 | auxin response factor 12 isoform X1 | ||||
| Swissprot | Q0J951 | 0.0 | ARFL_ORYSJ; Auxin response factor 12 | ||||
| Swissprot | Q258Y5 | 0.0 | ARFL_ORYSI; Auxin response factor 12 | ||||
| TrEMBL | A0A2K2CJD0 | 0.0 | A0A2K2CJD0_BRADI; Auxin response factor | ||||
| STRING | BRADI5G25767.1 | 0.0 | (Brachypodium distachyon) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G30330.2 | 0.0 | auxin response factor 6 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Brast09G253300.2.p |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



