 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Bv1_002900_doao.t1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Chenopodiaceae; Betoideae; Beta
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 665aa MW: 73185.2 Da PI: 7.075 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.6 | 9.1e-13 | 494 | 540 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
Bv1_002900_doao.t1 494 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAISYITELQ 540
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 8.9E-50 | 55 | 242 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.13 | 490 | 539 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 3.66E-18 | 493 | 552 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.84E-14 | 493 | 544 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 3.3E-18 | 494 | 554 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.9E-10 | 494 | 540 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 7.4E-17 | 496 | 545 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 665 aa Download sequence Send to blast |
MTNTELGMGG GVVWNEEDKA MIASILGTRA FDYLISSPFT AETSLVSFEN NENIQNKLSD 60 LVERPNASNF SWNYGFFWQI SRAKSGEVVL SWGDGYCREP REGEESEATR ILNFRLEDES 120 HQKIRKRVLQ KLNTLFGGSD DDNLACRLDR VTDMEMFFLV SMYFSFPRGE GGPGSCFVSG 180 KHVWESDLLR SKSDYCVRSF LAKSAGIQTV ILIPTEFGVV ELGSVRAIGE NHDVLKALKF 240 AFASPPVVRP KPTPVFPVVV HKIEEEDAPA LTPSPAPAPA PVRSPAPAPA RSPTPAPAPA 300 PAAVPASVPT TSPVPVSGPF SNVRLVDRGL DGVPKIFGQN LNTASRPPFR EKLAVKRMED 360 RHWDPYANGN KLSFAGQSPR NGLHGSNWAN IPGVKHATTT ELYSPQNTRL QEYVNGIRSE 420 YRLNQFHQQK PVQMEIDFSG ATSRPTVISQ PGSAESEHSD AEVSGRDDRA GPSDDRKPRK 480 RGRKPANGRE EPLNHVEAER QRREKLNQRF YALRAVVPNI SKMDKASLLG DAISYITELQ 540 KKLKDMEADR EKHNSTSKDT TSGTEANSVA DTQVQAPDVE VQAMHDEVVV RVSCPLDSHP 600 VARVIRAFKD SEINIVDSKL AAGTDTVFHT FVIKSQSSEQ LTKEKLLSAF SHESKPLQAP 660 SSTGG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 1e-26 | 488 | 550 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 1e-26 | 488 | 550 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 1e-26 | 488 | 550 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 1e-26 | 488 | 550 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 1e-26 | 488 | 550 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 1e-26 | 488 | 550 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 1e-26 | 488 | 550 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 1e-26 | 488 | 550 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 475 | 483 | RKPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that negatively regulates jasmonate (JA) signaling (PubMed:30610166). Negatively regulates JA-dependent response to wounding, JA-induced expression of defense genes, JA-dependent responses against herbivorous insects, and JA-dependent resistance against Botrytis cinerea infection (PubMed:30610166). Plays a positive role in resistance against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PubMed:30610166). {ECO:0000269|PubMed:30610166}. | |||||
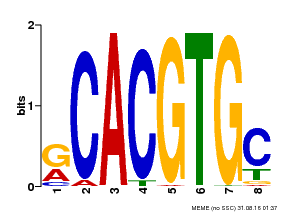
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding, feeding with herbivorous insects, infection with the fungal pathogen Botrytis cinerea and infection with the bacterial pathogen Pseudomonas syringae pv tomato DC3000. {ECO:0000269|PubMed:30610166}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010696336.1 | 0.0 | PREDICTED: transcription factor bHLH13 | ||||
| Swissprot | A0A3Q7ELQ2 | 0.0 | MTB1_SOLLC; Transcription factor MTB1 | ||||
| TrEMBL | A0A0K9QUK2 | 0.0 | A0A0K9QUK2_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010696336.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 1e-110 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



